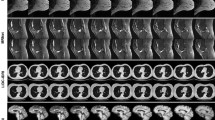
Abstract
Generative latent diffusion models have been established as state-of-the-art in data generation. One promising application is generation of realistic synthetic medical imaging data for open data sharing without compromising patient privacy. Despite the promise, the capacity of such models to memorize sensitive patient training data and synthesize samples showing high resemblance to training data samples is relatively unexplored. Here, we assess the memorization capacity of 3D latent diffusion models on photon-counting coronary computed tomography angiography and knee magnetic resonance imaging datasets. To detect potential memorization of training samples, we utilize self-supervised models based on contrastive learning. Our results suggest that such latent diffusion models indeed memorize training data, and there is a dire need for devising strategies to mitigate memorization.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Akbar, M.U., Wang, W., Eklund, A.: Beware of diffusion models for synthesizing medical images - a comparison with GANs in terms of memorizing brain tumor images (2023)
Bien, N., et al.: Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLOS Med. 15(11), 1–19 (2018). https://doi.org/10.1371/journal.pmed.1002699
Carlini, N., et al.: Extracting training data from diffusion models (2023)
Dorjsembe, Z., Odonchimed, S., Xiao, F.: Three-dimensional medical image synthesis with denoising diffusion probabilistic models. In: Medical Imaging with Deep Learning (2022)
Engelhardt, S., Sharan, L., Karck, M., Simone, R.D., Wolf, I.: Cross-domain conditional generative adversarial networks for stereoscopic hyperrealism in surgical training. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11768, pp. 155–163. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32254-0_18
Güngör, A., et al.: Adaptive diffusion priors for accelerated MRI reconstruction. Med. Image Anal. 102872 (2023). https://doi.org/10.1016/j.media.2023.102872
Ho, J., Jain, A., Abbeel, P.: Denoising diffusion probabilistic models. In: Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M.F., Lin, H. (eds.) Advances in Neural Information Processing Systems, vol. 33, pp. 6840–6851. Curran Associates, Inc. (2020)
Kazerouni, A., Aghdam, E.K., Heidari, M., Azad, R., Fayyaz, M., Hacihaliloglu, I., Merhof, D.: Diffusion models in medical imaging: a comprehensive survey. Med. Image Anal. 88, 102846 (2023). https://doi.org/10.1016/j.media.2023.102846
Khader, F., et al.: Denoising diffusion probabilistic models for 3D medical image generation. Sci. Rep. 13(1), 7303 (2023). https://doi.org/10.1038/s41598-023-34341-2
Özbey, M., Dalmaz, O., Dar, S.U., Bedel, H.A., Özturk, c., Güngör, A., Çukur, T.: Unsupervised medical image translation with adversarial diffusion models. IEEE Trans. Med. Imaging 1 (2023). https://doi.org/10.1109/TMI.2023.3290149
Pfeiffer, M., et al.: Generating large labeled data sets for laparoscopic image processing tasks using unpaired image-to-image translation. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11768, pp. 119–127. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32254-0_14
Pinaya, W.H.L., et al.: Brain imaging generation with latent diffusion models. In: Mukhopadhyay, A., Oksuz, I., Engelhardt, S., Zhu, D., Yuan, Y. (eds.) Deep Generative Models, DGM4MICCAI 2022, LNCS, vol. 13609, pp 117–126. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-18576-2_12
Rombach, R., Blattmann, A., Lorenz, D., Esser, P., Ommer, B.: High-resolution image synthesis with latent diffusion models. In: 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 10674–10685 (2022). https://doi.org/10.1109/CVPR52688.2022.01042
Schroff, F., Kalenichenko, D., Philbin, J.: FaceNet: a unified embedding for face recognition and clustering. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 2015
Somepalli, G., Singla, V., Goldblum, M., Geiping, J., Goldstein, T.: Diffusion art or digital forgery? investigating data replication in diffusion models. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 6048–6058, June 2023
Somepalli, G., Singla, V., Goldblum, M., Geiping, J., Goldstein, T.: Understanding and mitigating copying in diffusion models (2023)
Wolleb, J., Bieder, F., Sandkühler, R., Cattin, P.C.: Diffusion models for medical anomaly detection. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2022, pp. 35–45. Springer Nature Switzerland, Cham (2022). https://doi.org/10.1007/978-3-031-16452-1_4
Wolleb, J., Sandkühler, R., Bieder, F., Valmaggia, P., Cattin, P.C.: Diffusion models for implicit image segmentation ensembles. In: Konukoglu, E., Menze, B., Venkataraman, A., Baumgartner, C., Dou, Q., Albarqouni, S. (eds.) Proceedings of The 5th International Conference on Medical Imaging with Deep Learning. Proceedings of Machine Learning Research, vol. 172, pp. 1336–1348. PMLR, 06–08 July 2022
Yi, X., Walia, E., Babyn, P.: Generative adversarial network in medical imaging: a review. Med. Image Anal. 58, 101552 (2019). https://doi.org/10.1016/j.media.2019.101552
Acknowledgment
This work was supported through state funds approved by the State Parliament of Baden-Württemberg for the Innovation Campus Health + Life Science Alliance Heidelberg Mannheim, BMBF-SWAG Project 01KD2215D, and Informatics for life project through Klaus Tschira Foundation. The authors also gratefully acknowledge the data storage service SDS@hd supported by the Ministry of Science, Research and the Arts Baden-Württemberg (MWK) and the German Research Foundation (DFG) through grant INST 35/1314-1 FUGG and INST 35/1503-1 FUGG. The authors also acknowledge support by the state of Baden-Württemberg through bwHPC and the German Research Foundation (DFG) through grant INST 35/1597-1 FUGG.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Dar, S.U.H. et al. (2024). Investigating Data Memorization in 3D Latent Diffusion Models for Medical Image Synthesis. In: Mukhopadhyay, A., Oksuz, I., Engelhardt, S., Zhu, D., Yuan, Y. (eds) Deep Generative Models. MICCAI 2023. Lecture Notes in Computer Science, vol 14533. Springer, Cham. https://doi.org/10.1007/978-3-031-53767-7_6
Download citation
DOI: https://doi.org/10.1007/978-3-031-53767-7_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-53766-0
Online ISBN: 978-3-031-53767-7
eBook Packages: Computer ScienceComputer Science (R0)