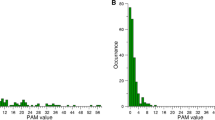
Abstract
Viruses are highly dependent on their hosts to carry out cellular mechanisms and cause productive infection. Thus, they undergo extensive adaptations to the host intracellular machinery, which occur over the evolution of the virus, and during the emergence of new viral strains with different properties. One aspect of viral adaptation is related to the efficiency of recruiting the host’s gene expression machinery and specifically the translation machinery. This process can be partially detected using measures of codon usage bias (CUB).
While previous studies in the field suggested that there is an adaptation of codons in the viral genome to the host, none of them studied these adaptations among the different strains of the same virus over time. Thus, in this study, we focused on the SARS-CoV-2 and demonstrated for the first time that the omicron strain has an increased codon usage adaptation to humans in the early gene ORF1ab compared to previous strains. In addition, our findings indicate that the observed differences in CUB scores were primarily attributed to non-synonymous mutations. This conclusion holds for additional human-infecting viruses.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Whitaker-Dowling, P., Youngner, J.S.: “VIRUS-HOST CELL INTERACTIONS,” in Encyclopedia of Virology, pp. 1957–1961. Elsevier (1999)
Lucas, M., Karrer, U., Lucas, A., Klenerman, P.: Viral escape mechanisms - escapology taught by viruses. Int. J. Exp. Pathol. 82(5), 269–286 (2008)
Goz, E., Zafrir, Z., Tuller, T.: Universal evolutionary selection for high dimensional silent patterns of information hidden in the redundancy of viral genetic code. Bioinformatics 34(19), 3241–3248 (2018)
Bahir, I., Fromer, M., Prat, Y., Linial, M.: Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Mol. Syst. Biol. 5(1), 311 (2009)
Crill, W.D., Wichman, H.A., Bull, J.J.: Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154(1), 27–37 (2000)
Sanjuán, R., Domingo-Calap, P.: Mechanisms of viral mutation. Cell. Mol. Life Sci. 73(23), 4433–4448 (2016)
Greenbaum, B.D., Levine, A.J., Bhanot, G., Rabadan, R.: Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS Pathog. 4(6), e1000079 (2008)
Elena, S.F., Sanjuán, R.: Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 79(18), 11555–11558 (2005)
Stern, A., Andino, R.: Viral Evolution. Viral Pathog. 233–240 (2016)
Holmes, E.C., Drummond, A.J.: The evolutionary genetics of viral emergence, pp. 51–66 (2007)
Duffy, S., Shackelton, L.A., Holmes, E.C.: Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9(4), 267–276 (2008)
Wong, E.H., Smith, D.K., Rabadan, R., Peiris, M., Poon, L.L.: Codon usage bias and the evolution of influenza A viruses. Codon usage biases of influenza virus. BMC Evol. Biol. 10, 253 (2010)
Khandia, R., et al.: Analysis of nipah virus codon usage and adaptation to hosts. Front. Microbiol. 10, 439603 (2019)
Cristina, J., Moreno, P., Moratorio, G., Musto, H.: Genome-wide analysis of codon usage bias in ebolavirus. Virus Res. 196, 87–93 (2015)
Biswas, K., et al.: Codon usage bias analysis of citrus tristeza virus: higher codon adaptation to citrus reticulata host. Viruses 11(4), 331 (2019)
Li, G., et al.: Evolutionary and genetic analysis of the VP2 gene of canine parvovirus. BMC Genomics 18(1), 534 (2017)
Cristina, J., Fajardo, A., Soñora, M., Moratorio, G., Musto, H.: A detailed comparative analysis of codon usage bias in Zika virus. Virus Res. 223, 147–152 (2016)
Moratorio, G., Iriarte, A., Moreno, P., Musto, H., Cristina, J.: A detailed comparative analysis on the overall codon usage patterns in West Nile virus. Infect. Genet. Evol. 14, 396–400 (2013)
Jenkins, G.M., Holmes, E.C.: The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 92(1), 1–7 (2003)
Belalov, I.S., Lukashev, A.N.: Causes and implications of codon usage bias in RNA viruses. PLoS ONE 8(2), e56642 (2013)
Parvez, M.K., Parveen, S.: Evolution and emergence of pathogenic viruses: past, present, and future. Intervirology 60(1–2), 1–7 (2017)
Pybus, O.G., Tatem, A.J., Lemey, P.: Virus evolution and transmission in an ever more connected world. Proc. R. Soc. B Biol. Sci. 282(1821), 20142878 (2015)
LaTourrette, K., Garcia-Ruiz, H.: Determinants of virus variation, evolution, and host adaptation. Pathogens 11(9), 1039 (2022)
Ojosnegros, S., Beerenwinkel, N.: Models of RNA virus evolution and their roles in vaccine design. Immunome Res. 6(Suppl 2), S5 (2010)
Hie, B., Zhong, E.D., Berger, B., Bryson, B.: Learning the language of viral evolution and escape. Science 371(6526), 284–288 (2021)
Marz, M., et al.: Challenges in RNA virus bioinformatics. Bioinformatics 30(13), 1793–1799 (2014)
Elena, S.F.: “Restrictions to RNA virus adaptation: an experimental approach”, Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 81(1–4), 135–142 (2002)
Hanna, R., Dalvi, S., Sălăgean, T., Pop, I.D., Bordea, I.R., Benedicenti, S.: Understanding COVID-19 pandemic: molecular mechanisms and potential therapeutic strategies. An evidence-based review. J. Inflamm. Res. 14, 13–56 (2021)
De Maio, N., Walker, C.R., Turakhia, Y., Lanfear, R., Corbett-Detig, R., Goldman, N.: Mutation rates and selection on synonymous mutations in SARS-CoV-2. Genome Biol. Evol. 13(5), evab087 (2021)
Magazine, N., Zhang, T., Wu, Y., McGee, M.C., Veggiani, G., Huang, W.: Mutations and evolution of the SARS-CoV-2 spike protein. Viruses 14(3), 640 (2022)
Nambou, K., Anakpa, M.: Deciphering the co-adaptation of codon usage between respiratory coronaviruses and their human host uncovers candidate therapeutics for COVID-19. Infect. Genet. Evol. 85, 104471 (2020)
Tao, K., et al.: The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 22(12), 757–773 (2021)
Planas, D., et al.: Reduced sensitivity of SARS-CoV-2 variant delta to antibody neutralization. Nature 596(7871), 276–280 (2021)
Hu, J., et al.: Increased immune escape of the new SARS-CoV-2 variant of concern omicron. Cell. Mol. Immunol. 19(2), 293–295 (2022)
Chavda, V., Bezbaruah, R., Deka, K., Nongrang, L., Kalita, T.: The delta and omicron variants of SARS-CoV-2: what we know so far. Vaccines 10(11), 1926 (2022)
Kumar, S., Thambiraja, T.S., Karuppanan, K., Subramaniam, G.: Omicron and delta variant of SARS-CoV-2: a comparative computational study of spike protein. J. Med. Virol. 94(4), 1641–1649 (2022)
Davidson, A.M., Wysocki, J., Batlle, D.: Interaction of SARS-CoV-2 and other coronavirus with ACE (Angiotensin-Converting Enzyme)-2 as their main receptor. Hypertension 76(5), 1339–1349 (2020)
Hamming, I., Timens, W., Bulthuis, M., Lely, A., Navis, G., van Goor, H.: Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203(2), 631–637 (2004)
Zhang, H., Penninger, J.M., Li, Y., Zhong, N., Slutsky, A.S.: Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 46(4), 586–590 (2020)
Lan, J., et al.: Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807), 215–220 (2020)
Peronace, C., et al.: The first identification in Italy of SARS-CoV-2 omicron BA. 4 harboring KSF141_del: a genomic comparison with omicron sub-variants. Biomedicines 10(8), 1839 (2022)
Chakraborty, C., Bhattacharya, M., Sharma, A.R., Dhama, K., Agoramoorthy, G.: A comprehensive analysis of the mutational landscape of the newly emerging omicron (B.1.1.529) variant and comparison of mutations with VOCs and VOIs. GeroScience 44(5), 2393–2425 (2022)
Plotkin, J.B., Dushoff, J.: Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc. Natl. Acad. Sci. 100(12), 7152–7157 (2003)
Koyama, T., Platt, D., Parida, L.: Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 98(7), 495–504 (2020)
Chatterjee, S., Bhattacharya, M., Nag, S., Dhama, K., Chakraborty, C.: A detailed overview of SARS-CoV-2 omicron: its sub-variants, mutations and pathophysiology, clinical characteristics, immunological landscape, immune escape, and therapies. Viruses 15(1), 167 (2023)
Hatcher, E.L., et al.: Virus variation resource – improved response to emergent viral outbreaks. Nucleic Acids Res. 45(D1), D482–D490 (2017)
Hodcroft, E.B.: CoVariants: SARS-CoV-2 Mutations and Variants of Interest (2021)
O’Leary, N.A., et al.: Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44(D1), D733–D745 (2016). https://doi.org/10.1007/978-3-319-21602-7_8
Alexaki, A., et al.: Codon and codon-pair usage tables (CoCoPUTs): facilitating genetic variation analyses and recombinant gene design. J. Mol. Biol. 431(13), 2434–2441 (2019)
Chan, P.P., Lowe, T.M.: GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37, D93–D97 (2009)
Chan, P.P., Lowe, T.M.: GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44(D1), D184–D189 (2016)
Sabi, R., Tuller, T.: Modelling the efficiency of codon–tRNA interactions based on codon usage bias. DNA Res. 21(5), 511–526 (2014)
Sharp, P.M., Li, W.-H.: The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15(3), 1281–1295 (1987)
Reis, M.D.: Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 32(17), 5036–5044 (2004)
Hernandez-Alias, X., Benisty, H., Schaefer, M.H., Serrano, L.: Translational efficiency across healthy and tumor tissues is proliferation-related. Mol. Syst. Biol. 16(3), e9275 (2020)
Pechmann, S., Frydman, J.: Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 20(2), 237–243 (2013)
Needleman, S.B., Wunsch, C.D.: A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48(3), 443–453 (1970)
Sievers, F., et al.: Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7(1), 539 (2011)
Davison, A.C., Hinkley, D.V.: Bootstrap Methods and their Application. Cambridge University Press, Cambridge (1997)
Bergman, S., Tuller, T.: Widespread non-modular overlapping codes in the coding regions*. Phys. Biol. 17(3), 031002 (2020)
Lauring, A.S., Frydman, J., Andino, R.: The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 11(5), 327–336 (2013)
Wang, R., Chen, J., Wei, G.-W.: Mechanisms of SARS-CoV-2 evolution revealing vaccine-resistant mutations in Europe and America. J. Phys. Chem. Lett. 12(49), 11850–11857 (2021)
Emam, M., Oweda, M., Antunes, A., El-Hadidi, M.: Positive selection as a key player for SARS-CoV-2 pathogenicity: insights into ORF1ab, S and E genes. Virus Res. 302, 198472 (2021)
V’kovski, P., Kratzel, A., Steiner, S., Stalder, H., Thiel, V.: Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19(3), 155–170 (2021)
Mioduser, O., Goz, E., Tuller, T.: Significant differences in terms of codon usage bias between bacteriophage early and late genes: a comparative genomics analysis. BMC Genomics 18(1), 866 (2017)
Manrubia, S., Lazaro, E.: Viral evolution. Phys. Life Rev. 3(2), 65–92 (2006)
Domingo, E., Holland, J.J.: RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51(1), 151–178 (1997)
Bull, R.A., et al.: Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog. 7(9), e1002243 (2011)
Domingo-Calap, P.: Viral evolution and Immune responses. J. Clin. Microbiol. Biochem. Technol. 5(2), 013–018 (2019)
Mordstein, C., et al.: Transcription, mRNA export, and immune evasion shape the codon usage of viruses. Genome Biol. Evol. 13(9), 1–14 (2021)
Nijhuis, M., Deeks, S., Boucher, C.: Implications of antiretroviral resistance on viral fitness. Curr. Opin. Infect. Dis. 14(1), 23–28 (2001)
Domingo, E., Menéndez-Arias, L., Holland, J.J.: RNA virus fitness. Rev. Med. Virol. 7(2), 87–96 (1997)
Gao, Y., et al.: Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368(6492), 779–782 (2020)
Daczkowski, C.M., Dzimianski, J.V., Clasman, J.R., Goodwin, O., Mesecar, A.D., Pegan, S.D.: Structural insights into the interaction of coronavirus papain-like proteases and interferon-stimulated gene product 15 from different species. J. Mol. Biol. 429(11), 1661–1683 (2017)
Acknowledgments
This research was funded by the DFG.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Davidson, A. et al. (2024). Evidence of Increased Adaptation of Omicron SARS-CoV-2 Codons to Humans. In: Scornavacca, C., Hernández-Rosales, M. (eds) Comparative Genomics. RECOMB-CG 2024. Lecture Notes in Computer Science(), vol 14616. Springer, Cham. https://doi.org/10.1007/978-3-031-58072-7_13
Download citation
DOI: https://doi.org/10.1007/978-3-031-58072-7_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-58071-0
Online ISBN: 978-3-031-58072-7
eBook Packages: Computer ScienceComputer Science (R0)