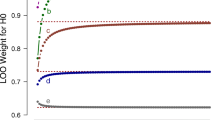
Abstract
Mutation validation (MV) is a recently proposed approach for model selection, garnering significant interest due to its unique characteristics and potential benefits compared to the widely used cross-validation (CV) method. In this study, we empirically compared MV and k-fold CV using benchmark and real-world datasets. By employing Bayesian tests, we compared generalization estimates yielding three posterior probabilities: practical equivalence, CV superiority, and MV superiority. We also evaluated the differences in the capacity of the selected models and computational efficiency. We found that both MV and CV select models with practically equivalent generalization performance across various machine learning algorithms and the majority of benchmark datasets. MV exhibited advantages in terms of selecting simpler models and lower computational costs. However, in some cases MV selected overly simplistic models leading to underfitting and showed instability in hyperparameter selection. These limitations of MV became more evident in the evaluation of a real-world neuroscientific task of predicting sex at birth using brain functional connectivity.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
xcpengine-container 1.0.1. https://pypi.org/project/xcpengine-container/
Barbiero, P., Squillero, G., Tonda, A.P.: Modeling generalization in machine learning: a methodological and computational study. CoRR abs/2006.15680 (2020)
Corani, G., Benavoli, A.: A Bayesian approach for comparing cross-validated algorithms on multiple data sets. Mach. Learn. 100, 285–304 (2015)
Corani, G., Benavoli, A., Demšar, J., Mangili, F., Zaffalon, M.: Statistical comparison of classifiers through Bayesian hierarchical modelling. Mach. Learn. 106(11), 1817–1837 (2017)
Dua, D., Graff, C.: UCI machine learning repository (2017). http://archive.ics.uci.edu/ml
Esteban, O., et al.: fMRIPrep: a robust preprocessing pipeline for functional MRI (2022)
Feldman, V., Frostig, R., Hardt, M.: The advantages of multiple classes for reducing overfitting from test set reuse. CoRR abs/1905.10360 (2019)
Guyon, I., Elisseeff, A.: An introduction to variable and feature selection. J. Mach. Learn. Res. 3, 1157–1182 (2003)
Hastie, T., Tibshirani, R., Friedman, J.: The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer Series in Statistics, Springer, New York (2009). https://doi.org/10.1007/978-0-387-21606-5
Kohavi, R.: A Study of Cross-validation and Bootstrap for Accuracy Estimation and Model Selection, pp. 1137–1143. IJCAI 1995, Morgan Kaufmann Publishers Inc., San Francisco, CA, USA (1995)
Kruschke, J.: Bayesian estimation supersedes the t test. J. Exp. Psychol. General 142, 573–603 (2012)
Mitchell, T.M.: Machine Learning. McGraw-hill, New York (1997)
Raschka, S.: Model evaluation, model selection, and algorithm selection in machine learning. CoRR abs/1811.12808 (2018)
Schaefer, A., et al.: Local-Global Parcellation of the Human Cerebral Cortex from Intrinsic Functional Connectivity MRI. Cerebral Cortex (2018)
Snoek, L., et al.: The Amsterdam open MRI collection, a set of multimodal MRI datasets for individual difference analyses. Sci. Data 8(1), 85 (2021)
Vanschoren, J., van Rijn, J.N., Bischl, B., Torgo, L.: OpenML: networked science in machine learning. SIGKDD Explor. 15(2), 49–60 (2013)
Weis, S., Patil, K.R., Hoffstaedter, F., Nostro, A., Yeo, B.T.T., Eickhoff, S.B.: Sex classification by resting state brain connectivity. Cereb. Cortex 30(2), 824–835 (2019)
Zhang, J.M., Harman, M., Guedj, B., Barr, E.T., Shawe-Taylor, J.: Model validation using mutated training labels: an exploratory study. Neurocomput. 539(C), 126116 (2023). https://doi.org/10.1016/j.neucom.2023.02.042
Acknowledgements
This work was partly supported by the Helmholtz Portfolio Theme “Supercomputing and Modelling for the Human Brain” and by the Max Planck School of Cognition supported by the Federal Ministry of Education and Research (BMBF) and the Max Planck Society (MPG).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Yu, J., Hamdan, S., Sasse, L., Morrison, A., Patil, K.R. (2024). Empirical Comparison Between Cross-Validation and Mutation-Validation in Model Selection. In: Miliou, I., Piatkowski, N., Papapetrou, P. (eds) Advances in Intelligent Data Analysis XXII. IDA 2024. Lecture Notes in Computer Science, vol 14642. Springer, Cham. https://doi.org/10.1007/978-3-031-58553-1_5
Download citation
DOI: https://doi.org/10.1007/978-3-031-58553-1_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-58555-5
Online ISBN: 978-3-031-58553-1
eBook Packages: Computer ScienceComputer Science (R0)