Abstract
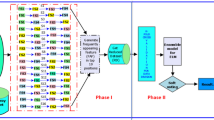
In this study, we present a novel approach to improve cancer classification using high-dimensional microarray data. The proposed method combines a hybrid filter and a genetic algorithm-based feature selection process, incorporating Chi-square and Recursive Feature Elimination (RFE) techniques to identify critical gene expressions for cancer classification. Experiments using diverse datasets have yielded significant results. In the Lung Cancer Dataset, Logistic Regression Analysis (LR) and Support Vector Machine (SVM) achieved remarkable accuracy rates of 97.56%, with a precision and recall of 98.0%, resulting in an F1-score of 97.0%. This highlights the effectiveness of the feature selection method in enhancing classification accuracy. In the Ovarian Cancer Dataset, Gradient Boosting emerged as the top-performing classifier, achieving an accuracy of 92.85% along with precision, recall, and F1-score values of 94.0%, 93.0%, and 92.0%, respectively. These results demonstrate the versatility of the proposed feature-selection approach. This demonstrates the adaptability of the proposed feature selection technique in improving classifier performance. In summary, the hybrid filter and genetic algorithm-based feature selection method, incorporating Chi-square and RFE, proved to be a valuable tool for enhancing cancer classification in high-dimensional microarray data. The consistently high accuracy, precision, recall, and F1-score across diverse cancer datasets underscore the effectiveness and versatility of the proposed approach, holding promise for the development of more accurate cancer classification models in the future.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others

References
Trevino, V., Falciani, F., Barrera-Saldaña, H.A.: DNA microarrays: a powerful genomic tool for biomedical and clinical research. Mol. Med. 13(9–10), 527–541 (2007). https://doi.org/10.2119/2006-00107.trevino
Cosma, G., Brown, D., Archer, M., Khan, M., Graham Pockley, A.: A survey on computational intelligence approaches for predictive modeling in prostate cancer. Expert Syst. Appl. 70, 1–19 (2017)
Lai, C.-M., Yeh, W.-C., Chang, C.-Y.: Gene selection using information gain and improved simplified swarm optimization. Neurocomputing 218, 331–338 (2016). https://doi.org/10.1016/j.neucom.2016.08.089
Singh, R.K., Sivabalakrishnan, M.: Feature selection of gene expression data for cancer classification: a review. Procedia Comput. Sci. 50, 52–57 (2023). https://doi.org/10.1016/j.procs.2015.04.060
Veerabhadrappa, M., Rangarajan, L.: Bi-level dimensionality reduc-tion methods using feature selection and feature extraction. Int. J. Comput. Appl. 4(2), 33–38 (2010). https://doi.org/10.5120/800-1137
Bennet, J., Ganaprakasam, C., Kumar, N.: A hybrid approach for gene selection and classification using support vector machine. Int. Arab J. Inf. Technol. 12(6A) (2015)
Jain, A., Zongker, D.: Feature selection: evaluation, application, and small sample performance. IEEE Trans. Pattern Anal. Mach. Intell. 19(2), 153–158 (1997). https://doi.org/10.1109/34.574797
Li, B.-Q., Hu, L.-L., Chen, L., Feng, K.-Y., Cai, Y.-D., Chou, K.-C.: Pre-diction of protein domain with mRMR feature selection and analysis. PLoS ONE 7(6), e39308 (2012). https://doi.org/10.1371/journal.pone.0039308
Alshamlan, H.M., Badr, G.H., Alohali, Y.A.: Genetic bee colony (GBC) algorithm: a new gene selection method for microarray cancer classification. Comput. Biol. Chem. 56, 49–60 (2015). https://doi.org/10.1016/j.compbiolchem.2015.03.001
García-Díaz, P., Sánchez-Berriel, I., Martínez-Rojas, J.A., Diez-Pascual, A.M.: Unsupervised feature selection algorithm for multiclass cancer classification of gene expression RNA-Seq data. Genomics 112(2), 1916–1925 (2020). https://doi.org/10.1016/j.ygeno.2019.11.004
Wu, J., Hicks, C.: Breast cancer type classification using machine learning. J. Personalized Med. 11(2), 61 (2021). https://doi.org/10.3390/jpm11020061
Chen, J.W., Dhahbi, J.: Lung adenocarcinoma and lung squamous cell carcinoma cancer classification, biomarker identification, and gene expression analysis using overlapping feature selection methods. Sci. Rep. 11(1) (2021). https://doi.org/10.1038/s41598-021-92725-8
Liu, S., Yao, W.: Prediction of lung cancer using gene expression and deep learning with KL divergence gene selection. BMC Bioinformatics 23(1) (2022). https://doi.org/10.1186/s12859-022-04689-9
Mahin, K.F., Robiuddin, M., Islam, M., Ashraf, S., Yeasmin, F., Shatabda, S.: PanClassif: improving pan cancer classification of single cell RNA-seq gene expression data using machine learning. Genomics 114(2), 110264 (2022). https://doi.org/10.1016/j.ygeno.2022.01.001
Gakii, C., Mireji, P.O., Rimiru, R.: Graph Based feature selection for reduction of dimensionality in next-generation RNA sequencing datasets. Algorithms 15(1), 21 (2022). https://doi.org/10.3390/a15010021
Li, Y., et al.: A comprehensive genomic pan-cancer classification using the cancer genome atlas gene expression data. BMC Genomics 18(1) (2017). https://doi.org/10.1186/s12864-017-3906-0
Zhang, Y., Deng, Q., Liang, W., Zou, X.: An efficient feature selection strategy based on multiple support vector machine technology with gene expression data. Biomed. Res. Int. 2018, 1–11 (2018). https://doi.org/10.1155/2018/7538204
Al Abir, F., Shovan, S.M., Hasan, M., Sayeed, A., Shin, J.: Biomarker identification by reversing the learning mechanism of an autoencoder and recursive feature elimination. Mol. Omics 18(7), 652–661 (2022). https://doi.org/10.1039/d1mo00467k
Zhu, Z., Ong, Y.S., Dash, M.: Markov blanket-embedded genetic algorithm for gene selection. Pattern Recogn. 49(11), 3236–3248 (2007)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Oyegbile, O., Saeed, F., Bamansoor, S. (2024). Hybrid Filter Feature Selection for Improving Cancer Classification in High-Dimensional Microarray Data. In: Saeed, F., Mohammed, F., Fazea, Y. (eds) Advances in Intelligent Computing Techniques and Applications. IRICT 2023. Lecture Notes on Data Engineering and Communications Technologies, vol 211. Springer, Cham. https://doi.org/10.1007/978-3-031-59707-7_26
Download citation
DOI: https://doi.org/10.1007/978-3-031-59707-7_26
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-59706-0
Online ISBN: 978-3-031-59707-7
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)