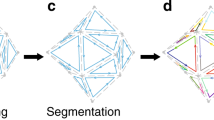
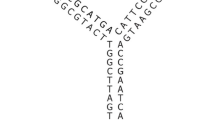
Abstract
In the area of DNA nanotechnology, approaches to composing wireframe nanostructures exclusively from short oligonucleotides, without a coordinating long scaffold strand, have been proposed by Goodman et al. (2004) and Wang et al. (2019). We present a general design method that extends these special cases to arbitrary wireframes, in the sense of graphs linearly embedded in 2D or 3D space. The method works in linear time in the size of the given wireframe model and is already available for use in the online design tool DNAforge. We also interpret the method in terms of topological graph embeddings, which opens up further research opportunities in developing this design approach.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Benson, E., Mohammed, A., Gardell, J., Masich, S., Czeizler, E., Orponen, P., Högberg, B.: DNA rendering of polyhedral meshes at the nanoscale. Nature 523(7561), 441–444 (2015). https://doi.org/10.1038/nature14586
Dey, S., et al.: DNA origami. Nat. Rev. Methods Primers 1(1), 13 (2021). https://doi.org/10.1038/s43586-020-00009-8
Dietz, H., Douglas, S.M., Shih, W.M.: Folding DNA into twisted and curved nanoscale shapes. Science 325(5941), 725 (2009). https://doi.org/10.1126/science.1174251
Douglas, S.M., Dietz, H., Liedl, T., Högberg, B., Graf, F., Shih, W.M.: Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459(7245), 414–418 (2009). https://doi.org/10.1038/nature08016
Ellingham, M.N., Ellis-Monaghan, J.A.: Bi-Eulerian embeddings of graphs and digraphs (2024). arXiv:2404.00325, https://doi.org/10.48550/arXiv.2404.00325
Ellis-Monaghan, J.A.: Transition polynomials, double covers, and biomolecular computing. Congr. Numer. 166, 181 (2004)
Ellis-Monaghan, J.A., McDowell, A., Moffatt, I., Pangborn, G.: DNA origami and the complexity of Eulerian circuits with turning costs. Nat. Comput. 14(3), 491–503 (2015). https://doi.org/10.1007/s11047-014-9457-2
Elonen, A., Wimbes, L., Mohammed, A., Orponen, P.: DNAforge: a design tool for nucleic acid wireframe nanostructures. Nucleic Acids Res. gkae367 (Online 15 May 2024). https://doi.org/10.1093/nar/gkae367
Fijavž, G., Pisanski, T., Rus, J.: Strong traces model of self-assembly polypeptide structures. MATCH Commun. Math. Comput. Chem. 71, 199–212 (2014). https://doi.org/10.48550/arXiv.1308.4024
Fleischner, H.: Eulerian Graphs and Related Topics. Part 1, vol. 1, Annals of Discrete Mathematics, vol. 45. North-Holland Publishing Co., Amsterdam (1990)
Furst, M.L., Gross, J.L., McGeoch, L.A.: Finding a maximum-genus graph imbedding. J. ACM (JACM) 35(3), 523–534 (1988). https://doi.org/10.1145/44483.44485
Goodman, R.P., Berry, R.M., Turberfield, A.J.: The single-step synthesis of a DNA tetrahedron. Chem. Commun. 40(12), 1372–1373 (2004). https://doi.org/10.1039/B402293A
Gross, J.L., Tucker, T.W.: Topological Graph Theory. Courier Corporation (2001)
Gross, J.L., Yellen, J., Zhang, P.: Handbook of Graph Theory, 2nd edn. CRC Press, Boca Raton (2014)
Jonoska, N., Saito, M.: Boundary components of thickened graphs. In: Jonoska, N., Seeman, N.C. (eds.) DNA Computing, pp. 70–81. Springer, Berlin Heidelberg (2002). https://doi.org/10.1007/3-540-48017-X_7
Jun, H., et al.: Rapid prototyping of arbitrary 2D and 3D wireframe DNA origami. Nucleic Acids Res. 49(18), 10265–10274 (2021). https://doi.org/10.1093/nar/gkab762
Jungnickel, D.: Graphs, Networks and Algorithm. Algorithms and Computation in Mathematics. Springer, Berlin, Heidelberg (2013). https://doi.org/10.1007/978-3-642-32278-5
Ke, Y., Ong, L.L., Shih, W.M., Yin, P.: Three-dimensional structures self-assembled from DNA bricks. Science 338(6111), 1177 (2012). https://doi.org/10.1126/science.1227268
Kuták, D., Poppleton, E., Miao, H., Šulc, P., Barišić, I.: Unified nanotechnology format: one way to store them all. Molecules 27(1), 63 (2022). https://doi.org/10.3390/molecules27010063
Lee, J.: Introduction to Topological Manifolds, vol. 202. Springer Science & Business Media, Cham (2010). https://doi.org/10.1007/978-1-4419-7940-7
Marchi, A.N., Saaem, I., Vogen, B.N., Brown, S., LaBean, T.H.: Toward larger DNA origami. Nano Lett. 14(10), 5740–5747 (2014). https://doi.org/10.1021/nl502626s
Ong, L.L., Hanikel, N., Yaghi, O.K., Grun, C., Strauss, M.T., Bron, P., Lai-Kee-Him, J., Schueder, F., Wang, B., Wang, P., Kishi, J.Y., Myhrvold, C., Zhu, A., Jungmann, R., Bellot, G., Ke, Y., Yin, P.: Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 552(7683), 72–77 (2017). https://doi.org/10.1038/nature24648
Orponen, P.: Design methods for 3D wireframe DNA nanostructures. Nat. Comput. 17(1), 147–160 (2018). https://doi.org/10.1007/s11047-017-9647-9
Rothemund, P.W.K.: Folding DNA to create nanoscale shapes and patterns. Nature 440(7082), 297–302 (2006). https://doi.org/10.1038/nature04586
Rovigatti, L., Šulc, P., Reguly, I.Z., Romano, F.: A comparison between parallelization approaches in molecular dynamics simulations on GPUs. J. Comput. Chem. 36(1), 1–8 (2015). https://doi.org/10.1002/jcc.23763
Seeman, N.C., Sleiman, H.F.: DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2017). https://doi.org/10.1038/natrevmats.2017.68
Seitz, S., Alava, M., Orponen, P.: Focused local search for random 3-satisfiability. J. Stat. Mech. Theory Exp. 2005(06), P06006 (2005). https://doi.org/10.1088/1742-5468/2005/06/P06006
Thomassen, C.: The graph genus problem is NP-complete. J. Algorithms 10(4), 568–576 (1989). https://doi.org/10.1016/0196-6774(89)90006-0
Veneziano, R., et al.: Designer nanoscale DNA assemblies programmed from the top down. Science (2016). https://doi.org/10.1126/science.aaf4388
Šulc, P., Romano, F., Ouldridge, T.E., Doye, J.P.K., Louis, A.A.: A nucleotide-level coarse-grained model of RNA. J. Chem. Phys. 140(23), 235102 (2014). https://doi.org/10.1063/1.4881424
Šulc, P., Romano, F., Ouldridge, T.E., Rovigatti, L., Doye, J.P.K., Louis, A.A.: Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 137(13), 135101 (2012). https://doi.org/10.1063/1.4754132
Wang, W., et al.: Complex wireframe DNA nanostructures from simple building blocks. Nat. Commun. 10(1), 1067 (2019). https://doi.org/10.1038/s41467-019-08647-7
Wei, B., Dai, M., Yin, P.: Complex shapes self-assembled from single-stranded DNA tiles. Nature 485(7400), 623–626 (2012). https://doi.org/10.1038/nature11075
Yin, P., Hariadi, R.F., Sahu, S., Choi, H.M.T., Park, S.H., LaBean, T.H., Reif, J.H.: Programming DNA tube circumferences. Science 321(5890), 824–826 (2008). https://doi.org/10.1126/science.1157312
Zhang, H., Chao, J., Pan, D., Liu, H., Huang, Q., Fan, C.: Folding super-sized DNA origami with scaffold strands from long-range PCR. Chem. Commun. 48(51), 6405–6407 (2012). https://doi.org/10.1039/c2cc32204h
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Elonen, A., Mohammed, A., Orponen, P. (2024). A General Design Method for Scaffold-Free DNA Wireframe Nanostructures. In: Cho, DJ., Kim, J. (eds) Unconventional Computation and Natural Computation. UCNC 2024. Lecture Notes in Computer Science, vol 14776. Springer, Cham. https://doi.org/10.1007/978-3-031-63742-1_13
Download citation
DOI: https://doi.org/10.1007/978-3-031-63742-1_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-63741-4
Online ISBN: 978-3-031-63742-1
eBook Packages: Computer ScienceComputer Science (R0)