Abstract
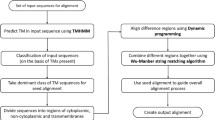
Transmembrane proteins (TMPs) are crucial to cell biology, making up about 30% of all proteins based on genomic data. Despite their importance, most of the available software for aligning protein sequences focuses on soluble proteins, leaving a gap in tools specifically designed for TMPs. Only a few methods target TMP alignment, with just a couple of the available to researchers. Considering that there are a few particular differences that ought to be taken into consideration aligning TMPs sequences, standard MSA methods are ineffective to align TMPs. In this paper, we present TM-MSAligner, a software tool designed to deal with the multiple sequence alignment of TMPs by using a multi-objective evolutionary algorithm. Our software include features such as transmembrane substitution matrix dynamically used according to the topology region, a high penalty to gap opening and extending, and two MSA quality scores, Sum-Of-Pairs with Topology Prediction and Aligned Segments, that can be optimized at the same time. This approach reduce the number of Transmembrane (TM) and non-Transmembrane (non-TM) broken regions and improve the TMP quality score. TM-MSAligner outputs the results in an HTML format, providing an interactive way for users to visualize and analyze the alignment. This feature allows for the easy identification of each topological region within the alignment, facilitating a quicker and more effective analysis process for researchers.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
MSABrowser: https://thekaplanlab.github.io/.
- 2.
TM-MSAligner: https://github.com/jMetal/TM-MSAligner.
References
Blum, C., Roli, A.: Metaheuristics in combinatorial optimization: overview and conceptual comparison. ACM Comput. Surv. 35(3), 268–308 (2003)
Coello Coello, C.A., Lamont, G.B., Van Veldhuizen, D.A.: Evolutionary Algorithms for Solving Multi-Objective Problems, 2nd edn. Springer, New York (2007). https://doi.org/10.1007/978-0-387-36797-2. iSBN 978-0-387-33254-3
Deb, K., Pratap, A., Agarwal, S., Meyarivan, T.: A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput. 6(2), 182–197 (2002)
Durillo, J.J., Nebro, A.J.: jMetal: a Java framework for multi-objective optimization. Adv. Eng. Softw. 42(10), 760–771 (2011)
Durillo, J.J., Nebro, A.J., Luna, F., Alba, E.: A study of master-slave approaches to parallelize NSGA-II. In: 2008 IEEE International Symposium on Parallel and Distributed Processing, pp. 1–8 (2008)
Hallgren, J., et al.: DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv (2022)
Henikoff, S., Henikoff, J.: Amino acid substitution matrices from protein blocks. Proc. Nat. Acad. Sci. 89(22), 10915–10919 (1992)
Nebro, A.J., Durillo, J.J., Vergne, M.: Redesigning the jMetal multi-objective optimization framework. In: Genetic and Evolutionary Computation Conference, pp. 1093–1100 (7 2015)
Ng, D.P., Poulsen, B.E., Deber, C.M.: Membrane protein misassembly in disease. Biochimica et Biophysica Acta (BBA) - Biomembranes 1818(4), 1115–1122 (2012). Protein Folding in Membranes
Ng, P.C., Henikoff, J.G., Henikoff, S.: PHAT: a transmembrane-specific substitution matrix. Bioinformatics 16, 760–766 (2000)
Pirovano, W., Abeln, S., Feenstra, K.A., Heringa, J.: Multiple alignment of transmembrane protein sequences. In: Structural Bioinformatics of Membrane Proteins, pp. 103–122. Springer, Vienna (2010). https://doi.org/10.1007/978-3-7091-0045-5_6
Thompson, J.D., Koehl, P., Ripp, R., Poch, O.: BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins Struct. Funct. Bioinf. 61(1), 127–136 (2005)
Torun, F.M., Bilgin, H.I., Kaplan, O.I.: MSABrowser: dynamic and fast visualization of sequence alignments, variations and annotations. Bioinf. Adv. 1(1), vbab009 (2021)
Wallin, E., Heijne, G.V.: Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7(4), 1029–1038 (1998)
Yin, H., Flynn, A.D.: Drugging membrane protein interactions. Ann. Rev. Biomed. Eng. 18(1), 51–76 (2016). pMID: 26863923
Zitzler, E., Laumanns, M., Thiele, L.: SPEA2: improving the strength pareto evolutionary algorithm. Technical report, 103, Swiss Federal Institute of Technology (ETH), Zurich, Switzerland (2001)
Acknowledgements
This work has been partially funded by the Spanish Ministry of Science and Innovation via Grant PID2020-112540RB-C41 (AEI/FEDER, UE) and by the Junta de Andalucía, Spain, under contract QUAL21 010UMA.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Cedeño-Muñoz, J., Zambrano-Vega, C., Nebro, A.J. (2024). TM-MSAligner: A Tool for Multiple Sequence Alignment of Transmembrane Proteins. In: Franco, L., de Mulatier, C., Paszynski, M., Krzhizhanovskaya, V.V., Dongarra, J.J., Sloot, P.M.A. (eds) Computational Science – ICCS 2024. ICCS 2024. Lecture Notes in Computer Science, vol 14835. Springer, Cham. https://doi.org/10.1007/978-3-031-63772-8_10
Download citation
DOI: https://doi.org/10.1007/978-3-031-63772-8_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-63771-1
Online ISBN: 978-3-031-63772-8
eBook Packages: Computer ScienceComputer Science (R0)