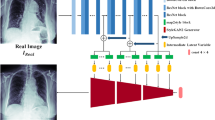
Abstract
Automated analytics in computational histopathology have shown significant progress in aiding pathologists through digital image analysis. However, developing a robust model based on supervised learning for histopathology images is challenging because of the scarcity of tumor-marked samples and unknown diseases. Unsupervised anomaly detection (UAD) methods that were mostly used in industrial inspection are thus proposed to facilitate efficient analytics. UAD only requires normal samples for training and largely reduces the burden of labeling. In this paper, we introduce a reconstruction-based UAD approach called PathUAD to improve representation learning based on adversarial learning and simulated anomalies. On the one hand, we mix up features extracted from normal images to build a smoother feature distribution and employ adversarial learning to enhance an autoencoder for image reconstruction. On the other hand, we simulate anomalous images by image deformation, and guide the autoencoder to catch global characteristics of normal images well. We demonstrate its effectiveness on a histopathology anomaly detection benchmark and show state-of-the-art performance.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Akcay, S., Atapour-Abarghouei, A., Breckon, T.P.: GANomaly: semi-supervised anomaly detection via adversarial training. In: Jawahar, C.V., Li, H., Mori, G., Schindler, K. (eds.) ACCV 2018. LNCS, vol. 11363, pp. 622–637. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-20893-6_39
Bao, J., Sun, H., Deng, H., He, Y., Zhang, Z., Li, X.: BMAD: benchmarks for medical anomaly detection. arXiv arXiv:2306.11876 (2023)
Bejnordi, B.E., et al.: The CAMELYON16 consortium: diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. J. Am. Med. Assoc. 318(22), 2199–2210 (2017)
Chang, Y.C., et al.: VCP maintains nuclear size by regulating the DNA damage-associated MDC1-p53-autophagy axis in Drosophila. Nat. Commun. 12(1), 4258 (2021)
Chen, L., You, Z., Zhang, N., Xi, J., Le, X.: UTRAD: anomaly detection and localization with U-transformer. Neural Netw. 147, 53–62 (2022)
Defard, T., Setkov, A., Loesch, A., Audigier, R.: PaDiM: a patch distribution modeling framework for anomaly detection and localization. In: Del Bimbo, A., et al. (eds.) ICPR 2021. LNCS, vol. 12664, pp. 475–489. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-68799-1_35
Deng, H., Li., X.: Anomaly detection via reverse distillation from one-class embedding. In: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9727–9736 (2022)
Goodfellow, I., et al.: Generative adversarial nets. In: Proceedings of Advances in Neural Information Processing Systems, vol. 27, pp. 2672–2680 (2014)
Gudovskiy, D., Ishizaka, S., Kozuka, K.: CFLOW-AD: real-time unsupervised anomaly detection with localization via conditional normalizing flows. In: Proceedings of IEEE Winter Conference on Applications of Computer Vision, pp. 1819–1828 (2022)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Ho, J., Jain, A., Abbeel, P.: Denoising diffusion probabilistic models. In: Proceedings of Advances in Neural Information Processing Systems, vol. 33, pp. 6840–6851 (2020)
Lee, S., Lee, S., Song, B.C.: CFA: coupled-hypersphere-based feature adaptation for target-oriented anomaly localization. IEEE Access 10, 78446–78454 (2022)
Li, B., Li, Y., Eliceiri, K.W.: Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. In: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 14313–14323 (2021)
Li, C.L., Yoon, K.S.J., Pfister, T.: CutPaste: self-supervised learning for anomaly detection and localization. In: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9659–9669 (2021)
Li, Y., Ping, W.: Cancer metastasis detection with neural conditional random field. arXiv arXiv:1806.07064 (2018)
Linmans, J., Raya, G., van der Laak, J., Litjens, G.: Diffusion models for out-of-distribution detection in digital pathology. Med. Image Anal. 93, 103088 (2024)
Liu, Z., Zhou, Y., Xu, Y., Wang, Z.: SimpleNet: a simple network for image anomaly detection and localization. In: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 20402–20411 (2023)
Lu, M.Y., Williamson, D.F.K., Chen, T.Y., Chen, R.J., Barbieri, M., Mahmood, F.: Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5(6), 555–570 (2021)
Mao, X., Li, Q., Xie, H., Lau, R.Y., Wang, Z., Smolley, S.P.: Least squares generative adversarial networks. In: Proceedings of IEEE/CVF International Conference on Computer Vision, pp. 2794–2802 (2017)
Meissen, F., Wiestler, B., Kaissis, G., Rueckert, D.: On the pitfalls of using the residual error as anomaly score. In: Proceedings of Medical Imaging with Deep Learning, pp. 914–928 (2022)
Milda, M.P., Eilertsen, G., Lundström, C.: Unsupervised anomaly detection in digital pathology using GANs. In: Proceedings of IEEE International Symposium on Biomedical Imaging, pp. 1878–1882 (2021)
Pinaya, W.H.L., et al.: Fast unsupervised brain anomaly detection and segmentation with diffusion models. In: Proceedings of Medical Image Computing and Computer Assisted Intervention, vol. 13438, pp. 705–714 (2022)
Roth, K., Pemula, L., Zepeda, J., Schölkopf, B., Brox, T., Gehler, P.: Towards total recall in industrial anomaly detection. In: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 14298–14308 (2022)
Rudolph, M., Wehrbein, T., Rosenhahn, B., Wandt, B.: Fully convolutional cross-scale-flows for image-based defect detection. In: Proceedings of IEEE Winter Conference on Applications of Computer Vision, pp. 1829–1838 (2022)
Ruff, L., et al.: Deep one-class classification. In: Proceedings of International Conference on Machine Learning, vol. 80, pp. 4393–4402 (2018)
Salehi, M., Sadjadi, N., Baselizadeh, S., Rohban, M.H., Rabiee, H.R.: Multiresolution knowledge distillation for anomaly detection. In: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 14897–14907 (2021)
Schlegl, T., Seeböck, P., Waldstein, S.M., Langs, G., Schmidt-Erfurth, U.: f-AnoGAN: fast unsupervised anomaly detection with generative adversarial networks. Med. Image Anal. 54, 30–44 (2019)
Shao, Z., et al.: TransMIL: transformer based correlated multiple instance learning for whole slide image classification. In: Proceedings of Advances in Neural Information Processing Systems, vol. 34, pp. 2136–2147 (2021)
Shvetsova, N., Bakker, B., Fedulova, I., Schulz, H., Dylov, D.V.: Anomaly detection in medical imaging with deep perceptual autoencoders. IEEE Access 9, 118571–118583 (2021)
Wang, T.C., Liu, M.Y., Zhu, J.Y., Tao, A., Kautz, J., Catanzaro, B.: High-resolution image synthesis and semantic manipulation with conditional GANs. In: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 8798–8807 (2018)
Wang, Z., Bovik, A., Sheikh, H., Simoncelli, E.: Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13(4), 600–612 (2004)
Yamada, S., Hotta, K.: Reconstruction student with attention for student-teacher pyramid matching. arXiv arXiv:2111.15376 (2021)
Zavrtanik, V., Kristan, M., Skočaj, D.: DrÆm - a discriminatively trained reconstruction embedding for surface anomaly detection. In: Proceedings of IEEE/CVF International Conference on Computer Vision, pp. 8310–8319 (2021)
Zhang, X., Li, N., Li, J., Dai, T., Jiang, Y., Xia, S.T.: Unsupervised surface anomaly detection with diffusion probabilistic model. In: Proceedings of IEEE/CVF International Conference on Computer Vision, pp. 6759–6768 (2023)
Štepec, D., Skočaj, D.: Unsupervised detection of cancerous regions in histology imagery using image-to-image translation. In: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, pp. 3780–3787 (2021)
Acknowledgement
This work was funded in part by Qualcomm through a Taiwan University Research Collaboration Project and in part by the National Science and Technology Council, Taiwan, under grants 113-2425-H-006-007, 112-2634-F-006-002, 112-2221-E-006-136-MY3, 112-2622-E-006-021, and 110-2221-E-006-127-MY3.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Lai, YC., Chu, WT. (2024). Unsupervised Anomaly Detection on Histopathology Images Using Adversarial Learning and Simulated Anomaly. In: Yap, M.H., Kendrick, C., Behera, A., Cootes, T., Zwiggelaar, R. (eds) Medical Image Understanding and Analysis. MIUA 2024. Lecture Notes in Computer Science, vol 14859. Springer, Cham. https://doi.org/10.1007/978-3-031-66955-2_25
Download citation
DOI: https://doi.org/10.1007/978-3-031-66955-2_25
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-66954-5
Online ISBN: 978-3-031-66955-2
eBook Packages: Computer ScienceComputer Science (R0)