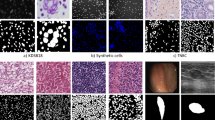
Abstract
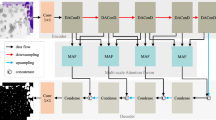
Accurate segmentation of cancer cell nuclei in medical images is fundamental for clinicians to diagnose and evaluate tumors, enabling scientists to explore genetic variants and epigenetic changes in cancer cells and reveal the complex mechanisms of tumorigenesis. Fine cell nucleus segmentation not only improves the accuracy of cancer diagnosis, but is also an important tool for personalized treatment and advances in cancer research. Currently, the representatives of image segmentation models in the field of medical image analysis, various improved versions of U-Net, exhibit suboptimal performance on certain datasets. The challenges arise from the non-ideal shooting conditions prevalent in medical imaging, characterized by low-light environments and low-pixel slices. Compounding the issue is the diminutive size of cell nuclei relative to the overall image, coupled with their potentially abundant and densely distributed nature within the frame. Based on these difficult thinking on, a method is used SPD convolution to construct a new multi-feature fusion layer, and combined with the SimAM attention mechanism to improve the current YOLOv8 model. YOLO-TL achieves a Dice score of 0.852 and a Jaccard coefficient of 0.7804, where the Dice score is at least 2% points or more higher than a host of improved U-Net-based models.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Qin, J., Puckett, L., Qian, X.: Image based fractal analysis for detection of cancer cells. In: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Korea (South), pp. 1482–1486 (2020). https://doi.org/10.1109/BIBM49941.2020.9313176
Zhao, Z., Wang, H., Zhang, Y., Zheng, H., Zhang, S., Chen, D.Z.: A coarse-to-fine data generation method for 2D and 3D cell nucleus segmentation. In: 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Rochester, MN, USA, pp. 41–46 (2020). https://doi.org/10.1109/CBMS49503.2020.00016
Saednia, K., Tran, W.T., Sadeghi-Naini, A.: A cascaded deep learning framework for segmentation of nuclei in digital histology images. In: 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, Scotland, United Kingdom, pp. 4764–4767 (2022). https://doi.org/10.1109/EMBC48229.2022.9871996
Balwant, M.K.: A review on convolutional neural networks for brain tumor segmentation: methods, datasets, libraries, and future directions. IRBM 43(6), 521–537 (2022). https://doi.org/10.1016/j.irbm.2022.05.002
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. arXiv:1505.04597 [cs.CV] (2015). https://doi.org/10.48550/arXiv.1505.04597
Wu, Y., Wu, J., Jin, S., Cao, L., Jin, G.: Dense-U-Net: dense encoder-decoder network for holographic imaging of 3D particle fields. Opt. Commun. 493, 126970 (2021). https://doi.org/10.1016/j.optcom.2021.126970
BBBC038v1 Image Set. Available from the Broad Bioimage Benchmark Collection. https://bbbc.broadinstitute.org/BBBC038
Naylor, P.J., Walter, T., Laé, M., Reyal, F.: Segmentation of Nuclei in Histopathology Images by deep regression of the distance map (1.0) [Data set]. In: Zenodo (2018). https://doi.org/10.5281/zenodo.1175282
Oktay, O., Schlemper, J., Le Folgoc, L., et al.: Attention U-Net: learning where to look for the pancreas. arXiv:1804.03999 [cs.CV] (2018). https://doi.org/10.48550/arXiv.1804.03999
Chen, L.C., Papandreou, G., Schroff, F., Adam, H.: Rethinking atrous convolution for semantic image segmentation. arXiv:1706.05587 [cs.CV] (2017). https://doi.org/10.48550/arXiv.1706.05587
Sun, Y.N., Yen, G.G., Zhang, M.J.: Evolutionary deep neural architecture search: fundamentals, methods, and recent advances, 8 November 2022. 978-3-031-16868-0, https://doi.org/10.1007/978-3-031-16868-0
Liu, M., Liu, H., He, X., et al.: Research advances on non-line-of-sight imaging technology. J. Shanghai Jiaotong Univ. (Sci.) (2024). https://doi.org/10.1007/s12204-023-2686-8
Steiner, A., Kolesnikov, A., Zhai, X., Wightman, R., Uszkoreit, J., Beyer, L.: How to train your ViT? Data, augmentation, and regularization in vision transformers. arXiv:2106.10270 [cs.CV] (2022). https://doi.org/10.48550/arXiv.2106.10270
Redmon, J., Farhadi, A.: YOLOv3: an incremental improvement. arXiv preprint arXiv:1804.02767 (2018). https://doi.org/10.48550/arXiv.1804.02767
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. (TPAMI) (2018). https://doi.org/10.1109/TPAMI.2017.2699184
He, K., Gkioxari, G., Dollár, P., Girshick, R.: Mask R-CNN. In: 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, pp. 2980–2988 (2017). https://doi.org/10.1109/ICCV.2017.322
Lin, T.Y., Dollár, P., Girshick, R., He, K., Hariharan, B., Belongie, S.: Feature pyramid networks for object detection. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, pp. 936–944 (2017). https://doi.org/10.1109/CVPR.2017.106
Dai, J., He, K., Sun, J.: Instance-aware semantic segmentation via multi-task network cascades. arXiv:1512.04412 [cs.CV] (2015). https://doi.org/10.48550/arXiv.1512.04412
Chen, L.C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) Computer Vision – ECCV 2018. ECCV 2018. LNCS, vol. 11211, pp. 833–851. Springer, Cham (2018). https://doi.org/10.48550/arXiv.1802.02611, https://doi.org/10.1007/978-3-030-01234-2_49
Jiang, H., Wang, J., Yuan, Z., Wu, Y., Zheng, N., Li, S.: Salient object detection: a discriminative regional feature integration approach. In: 2013 IEEE Conference on Computer Vision and Pattern Recognition, Portland, OR, USA, pp. 2083–2090 (2013). https://doi.org/10.1109/CVPR.2013.271
Ultralytics YOLOv8 Docs. https://docs.ultralytics.com/
Ding, X., Zhang, X., Zhou, Y., Han, J., Ding, G., Sun, J.: Scaling up your Kernels to 31 \(\times \) 31: revisiting large Kernel design in CNNs. In: CVPR (2022). https://doi.org/10.48550/arXiv.2203.06717
Dai, J., et al.: Deformable convolutional networks. In: Computer Vision and Pattern Recognition (cs.CV) (2017). https://doi.org/10.48550/arXiv.1703.06211
Qi, Y., He, Y., Qi, X., Zhang, Y., Yang, G.: Dynamic Snake Convolution based on topological geometric constraints for tubular structure segmentation. In: ICCV (2023). https://doi.org/10.48550/arXiv.2307.08388
Raja, S., Tie, L.: No more strided convolutions or pooling: a new CNN building block for low-resolution images and small objects. In: European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases (ECML PKDD) (2022). https://doi.org/10.48550/arXiv.2208.03641
Qiao, S., Chen, L.-C., Yuille, A.: DetectoRS: detecting objects with recursive feature pyramid and switchable atrous convolution. In: Computer Vision and Pattern Recognition (cs.CV) (2020). https://doi.org/10.48550/arXiv.2006.02334
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Computer Vision and Pattern Recognition (cs.CV) (2016). https://doi.org/10.48550/arXiv.1512.03385
Yang, L., Zhang, R.-Y., Li, L., Xie, X.: SimAM: a simple, parameter-free attention module for convolutional neural networks. In: Proceedings of the 38th International Conference on Machine Learning, PMLR, vol. 139, pp. 11863–11874 (2021). https://proceedings.mlr.press/v139/yang21o.html
Sebastian, R.: An overview of gradient descent optimization algorithms. In: Machine Learning (cs.LG) (2016). https://doi.org/10.48550/arXiv.1609.04747
Gabella, M.: Topology of learning in feedforward neural networks. IEEE Trans. Neural Netw. Learn. Syst. 32(8), 3588–3592 (2021). https://doi.org/10.1109/TNNLS.2020.3015790
Hafeez, M.A., Ul-Hasan, A., Shafait, F.: Incremental learning of object detector with limited training data. In: 2021 Digital Image Computing: Techniques and Applications (DICTA), Gold Coast, Australia, pp. 01–08 (2021). https://doi.org/10.1109/DICTA52665.2021.9647245
Fausto, M., Nassir, N., Seyed-Ahmad, A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: Computer Vision and Pattern Recognition (cs.CV) (2016). https://doi.org/10.48550/arXiv.1606.04797
Jeroen, B., et al.: Optimizing the dice score and Jaccard index for medical image segmentation: theory & practice. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11765. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32245-8_11
Dai, Y., Gieseke, F., Oehmcke, S., Wu, Y., Barnard, K.: Attentional feature fusion. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), pp. 3560–3569 (2021). https://doi.org/10.48550/arXiv.2009.14082
Liu, Z., Wei, J., Li, R., Zhou, J.: SFusion: self-attention based N-to-One Multimodal Fusion Block. In: Greenspan, H., et al. (eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. MICCAI 2023. LNCS, vol. 14221, pp. 159–169. Springer, Cham (2023). https://doi.org/10.1007/978-3-031-43895-0_15, https://doi.org/10.48550/arXiv.2208.12776
Acknowledgement
This work was supported in part by the National Natural Science Foundation of China (62102136) and 2022 University Teacher Characteristic Innovation Research Project of Foshan Education Bureau (2022DZXX04).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Li, Y., Duan, Y., Duan, L., Xiang, W., Wu, Q. (2024). YOLO-TL: A Tiny Object Segmentation Framework for Low Quality Medical Images. In: Yap, M.H., Kendrick, C., Behera, A., Cootes, T., Zwiggelaar, R. (eds) Medical Image Understanding and Analysis. MIUA 2024. Lecture Notes in Computer Science, vol 14860. Springer, Cham. https://doi.org/10.1007/978-3-031-66958-3_11
Download citation
DOI: https://doi.org/10.1007/978-3-031-66958-3_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-66957-6
Online ISBN: 978-3-031-66958-3
eBook Packages: Computer ScienceComputer Science (R0)