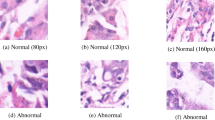
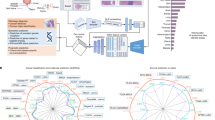
Abstract
Accurate analysis of histopathology images is a key step in cancer diagnosis and treatment planning. Specialized computational models for histopathology image analysis may be needed to better capture the unique characteristics of histopathological images that could be missed by models pretrained on generic data, such as ImageNet. We employed an incremental learning approach to enhance the performance of foundation models using the EfficientNet B0 architecture. Our study comprised three key experiments. First, we established a baseline accuracy of 84.4% by training the model on 50% of our dataset. Second, we investigated the impact of retraining different numbers of top blocks during incremental learning phases, finding that retraining three to six blocks provided the most significant accuracy improvements. Third, we analyzed the trade-off between accuracy improvement and training time, determining that retraining three to six blocks offered the best balance between performance gains and computational efficiency. Our results demonstrate the effectiveness of specialized models in capturing the details specific of histopathological images. Despite computational limitations, our findings underscore the importance of tailored histopathological models.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Aresta, G., et al.: BACH: grand challenge on breast cancer histology images. Med. Image Anal. 56, 122–139 (2019)
Arooj, S., Zubair, M., Khan, M.F., Alissa, K., Khan, M.A., Mosavi, A.: Breast cancer detection and classification empowered with transfer learning. Front. Public Health 10, 924432 (2022)
Arunachalam, H.B., et al.: Viable and necrotic tumor assessment from WSI of osteosarcoma using ML and DL models. PLoS ONE 14(4), e0210706 (2019)
Kather, J.N., Halama, N., Marx, A.: 100,000 histological images of human colorectal cancer and healthy tissue. Zenodo10 5281 (2018)
Kumar, A., Vishwakarma, A., Bajaj, V.: ML3CNet: non-local means-assisted automatic framework for lung cancer subtypes classification using histopathological images. Comput. Methods Programs Biomed. 251, 108207 (2024)
Mahmud, M.I., Mamun, M., Abdelgawad, A.: A deep analysis of transfer learning based breast cancer detection using histopathology images. In: 2023 10th International Conference on Signal Processing and Integrated Networks (SPIN), pp. 198–204. IEEE (2023)
Nasir, M.U., Khan, S., Mehmood, S., Khan, M.A., Rahman, A.U., Hwang, S.O.: IoMT-based osteosarcoma cancer detection in histopathology images using transfer learning empowered with blockchain, fog computing, and edge computing. Sensors 22(14), 5444 (2022)
Paszke, A., et al.: PyTorch: an imperative style, high-performance deep learning library. In: Proceedings of the 33rd International Conference on Neural Information Processing Systems (NeurIPS) (2019)
Pedregosa, F., et al.: Scikit-learn: ML in Python. JMLR 12, 2825–2830 (2011)
Silva-Rodríguez, J., Colomer, A., Sales, M.A., Molina, R., Naranjo, V.: Going deeper through the Gleason scoring scale: an automatic end-to-end system for histology prostate grading and cribriform pattern detection. Comput. Methods Programs Biomed. 195, 105637 (2020)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2016)
Tan, M., Le, Q.V.: EfficientNet: rethinking model scaling for convolutional neural networks. In: Proceedings of the 36th International Conference on Machine Learning (ICML), pp. 6105–6114 (2019)
Wu, Y., et al.: Recent advances of deep learning for computational histopathology: principles and applications. Cancers 14(5), 1199 (2022)
Yadav, A., Ahmed, F., Daescu, O., Gedik, R., Coskunuzer, B.: Histopathological cancer detection with topological signatures. In: 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 1610–1619. IEEE (2023)
Yadav, A., Daescu, O., Leavey, P., Rudzinski, E.: Machine learning for rhabdomyosarcoma whole slide images sub-type classification. In: Proceedings of the 16th International Conference on PErvasive Technologies Related to Assistive Environments, pp. 192–196 (2023)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Yadav, A., Daescu, O. (2024). Optimizing Foundation Models for Histopathology: A Continual Learning Approach to Cancer Detection. In: Chen, H., Zhou, Y., Xu, D., Vardhanabhuti, V.V. (eds) Trustworthy Artificial Intelligence for Healthcare. TAI4H 2024. Lecture Notes in Computer Science, vol 14812. Springer, Cham. https://doi.org/10.1007/978-3-031-67751-9_12
Download citation
DOI: https://doi.org/10.1007/978-3-031-67751-9_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-67750-2
Online ISBN: 978-3-031-67751-9
eBook Packages: Computer ScienceComputer Science (R0)