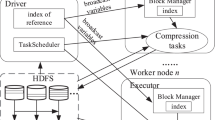
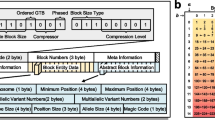
Abstract
Nowadays, Genomics has gained relevance since it allows preventing, diagnosing and treating diseases in a personalized way. The reduction in sequencing time and cost has increased the demand and, thus, the amount of genomic data that must be stored or transferred. Consequently, it becomes necessary to develop genome compression algorithms that help to reduce storage usage without consuming too much time. This is now possible thanks to modern multicore machines. This paper improves MtHRCM, a multi-threaded compression algorithm for large collections of genomes, by reducing its sequential component in order to enhance performance and scalability. Experimental results show that our optimized version is faster than MtHRCM and achieves the same compression ratio. Also, they reveal that this new version scales well when increasing the number of threads/cores for smaller test collections, while the high amount of simultaneous I/O requests to disk limits the scalability for larger test collections.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
NHS England: Health Education England’s Genomics Education Programme: what is genomics?. https://www.genomicseducation.hee.nhs.uk/education/core-concepts/what-is-genomics/
National Research Council: Mapping and Sequencing the Human Genome, p. 1988. The National Academies Press, Washington, DC (1988)
Drew, L.: Pharmacogenetics: the right drug for you. Nature 537, S60–S62 (2016)
Wetterstrand KA.: DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). www.genome.gov/sequencingcostsdata
National Human Genome Research Institute: Frequently Asked Questions and Resources. https://www.genome.gov/Clinical-Research/Secondary-Genomics-Findings-Service/FAQ-Resources
National Human Genome Research Institute: Genomic Data Science. https://www.genome.gov/about-genomics/fact-sheets/Genomic-Data-Science
Stephens, Z.D., et al.: Big Data: Astronomical or Genomical? PLoS Biol. 13(7), e1002195 (2015)
Kredens, K.V., et al.: Vertical lossless genomic data compression tools for assembled genomes: a systematic literature review. PLOS One 15(5), e0232942 (2020)
Hosseini, M., et al.: A survey on data compression methods for biological sequences. Information 7(4), 56 (2016)
Wandelt, S., et al.: Trends in genome compression. Curr Bioinf. 9(3), 315–326 (2013)
Deorowicz, S., et al.: GDC 2: compression of large collections of genomes. Sci. Rep. 5, 11565 (2015)
Yao H,. et al.: HRCM: an efficient hybrid referential compression method for genomic big data. BioMed. Res. Int. 2019, Article ID 3108950 (2019)
Whitehoyse D., Rapley R.: Chapter 5: Introductory bioinformatics. In: Genomics and Clinical Diagnostics. Royal Society of Chemistry (2019)
Gebank: NIH genetic sequence database. https://www.ncbi.nlm.nih.gov/genbank/
Wheeler, D., Bhagwat, M.: BLAST QuickStart: example-driven web-based BLAST tutorial. Methods Mol. Biol. 395, 149–176 (2007)
Yao, H., et al.: Parallel compression for large collections of genomes. Concurr. Comput. Pract. Exper. 34(2), e6339 (2021)
The International Genome Sample Resource (IGSR). https://www.internationalgenome.org/
UCSC Genome Browser Group: University of California, Santa Cruz. http://genome.ucsc.edu
Ahn, S.M., et al.: The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 19(9), 1622–1629 (2009)
KOBIC: Korea Bioinformation Center. ftp://ftp.kobic.kr/pub/KOBIC-KoreanGenome/
The National Center for Biotechnology Information, U.S.: Genome assembly HuRef. https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000002125.1/
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2025 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Sanz, V., Pousa, A., Naiouf, M., De Giusti, A. (2025). Fast Genomic Data Compression on Multicore Machines. In: Naiouf, M., De Giusti, L., Chichizola, F., Libutti, L. (eds) Cloud Computing, Big Data and Emerging Topics. JCC-BD&ET 2024. Communications in Computer and Information Science, vol 2189. Springer, Cham. https://doi.org/10.1007/978-3-031-70807-7_1
Download citation
DOI: https://doi.org/10.1007/978-3-031-70807-7_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-70806-0
Online ISBN: 978-3-031-70807-7
eBook Packages: Computer ScienceComputer Science (R0)