Abstract
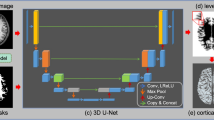
Cortical surface reconstruction typically relies on high-quality 3D brain MRI to establish the structure of cortex, playing a pivotal role in unveiling neurodevelopmental patterns. However, clinical challenges emerge due to elevated costs and prolonged acquisition times, often resulting in low-quality 2D brain MRI. To optimize the utilization of clinical data for cerebral cortex analysis, we propose a two-stage method for cortical surface reconstruction from 2D brain MRI images. The first stage employs segmentation-constrained MRI super-resolution, concatenating the super-resolution (SR) model and cortical ribbon segmentation model to emphasize cortical regions in the 3D images generated from 2D inputs. In the second stage, two encoders extract features from the original and super-resolution images. Through a shared decoder and the mask-swap module with multi-process training strategy, cortical surface reconstruction is achieved by mapping features from both the original and super-resolution images to a unified latent space. Experiments on the developing Human Connectome Project (dHCP) dataset demonstrate a significant improvement in geometric accuracy over the leading-SR based cortical surface reconstruction methods, facilitating precise cortical surface reconstruction from 2D images. The code is open-sourced at: https://github.com/SCUT-Xinlab/CSR-from-2D-MRI.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Jelena Bozek, Antonios Makropoulos, Andreas Schuh, Sean Fitzgibbon, Robert Wright, Matthew F Glasser, Timothy S Coalson, Jonathan O’Muircheartaigh, Jana Hutter, Anthony N Price, et al. Construction of a neonatal cortical surface atlas using multimodal surface matching in the developing human connectome project. NeuroImage, 179:11–29, 2018.
Rodrigo Santa Cruz, Leo Lebrat, Pierrick Bourgeat, Clinton Fookes, Jurgen Fripp, and Olivier Salvado. Deepcsr: A 3d deep learning approach for cortical surface reconstruction. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pages 806–815, 2021.
Anders M Dale, Bruce Fischl, and Martin I Sereno. Cortical surface-based analysis: I. segmentation and surface reconstruction. Neuroimage, 9(2):179–194, 1999.
Bruce Fischl. Freesurfer. Neuroimage, 62(2):774–781, 2012.
Bruce Fischl, Martin I Sereno, and Anders M Dale. Cortical surface-based analysis: Ii: inflation, flattening, and a surface-based coordinate system. Neuroimage, 9(2):195–207, 1999.
Emer J Hughes, Tobias Winchman, Francesco Padormo, Rui Teixeira, Julia Wurie, Maryanne Sharma, Matthew Fox, Jana Hutter, Lucilio Cordero-Grande, Anthony N Price, et al. A dedicated neonatal brain imaging system. Magnetic resonance in medicine, 78(2):794–804, 2017.
Leo Lebrat, Rodrigo Santa Cruz, Frederic de Gournay, Darren Fu, Pierrick Bourgeat, Jurgen Fripp, Clinton Fookes, and Olivier Salvado. Corticalflow: a diffeomorphic mesh transformer network for cortical surface reconstruction. Advances in Neural Information Processing Systems, 34:29491–29505, 2021.
Junnan Li, Dongxu Li, Silvio Savarese, and Steven Hoi. Blip-2: Bootstrapping language-image pre-training with frozen image encoders and large language models. arXiv preprint arXiv:2301.12597, 2023.
Junnan Li, Dongxu Li, Caiming Xiong, and Steven Hoi. Blip: Bootstrapping language-image pre-training for unified vision-language understanding and generation. In International Conference on Machine Learning, pages 12888–12900. PMLR, 2022.
Zhiyang Lu, Zheng Li, Jun Wang, Jun Shi, and Dinggang Shen. Two-stage self-supervised cycle-consistency network for reconstruction of thin-slice mr images. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part VI 24, pages 3–12. Springer, 2021.
Zhiyang Lu, Jian Wang, Zheng Li, Shihui Ying, Jun Wang, Jun Shi, and Dinggang Shen. Two-stage self-supervised cycle-consistency transformer network for reducing slice gap in mr images. IEEE Journal of Biomedical and Health Informatics, 2023.
Qiang Ma, Liu Li, Vanessa Kyriakopoulou, Joseph V Hajnal, Emma C Robinson, Bernhard Kainz, and Daniel Rueckert. Conditional temporal attention networks for neonatal cortical surface reconstruction. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 312–322. Springer, 2023.
Qiang Ma, Liu Li, Emma C Robinson, Bernhard Kainz, Daniel Rueckert, and Amir Alansary. Cortexode: Learning cortical surface reconstruction by neural odes. IEEE Transactions on Medical Imaging, 42(2):430–443, 2022.
Qiang Ma, Emma C Robinson, Bernhard Kainz, Daniel Rueckert, and Amir Alansary. Pialnn: a fast deep learning framework for cortical pial surface reconstruction. In Machine Learning in Clinical Neuroimaging: 4th International Workshop, MLCN 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, September 27, 2021, Proceedings 4, pages 73–81. Springer, 2021.
Soumi Maiti, Yifan Peng, Takaaki Saeki, and Shinji Watanabe. Speechlmscore: Evaluating speech generation using speech language model. In ICASSP 2023-2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pages 1–5. IEEE, 2023.
Antonios Makropoulos, Emma C Robinson, Andreas Schuh, Robert Wright, Sean Fitzgibbon, Jelena Bozek, Serena J Counsell, Johannes Steinweg, Katy Vecchiato, Jonathan Passerat-Palmbach, et al. The developing human connectome project: A minimal processing pipeline for neonatal cortical surface reconstruction. Neuroimage, 173:88–112, 2018.
Alec Radford, Jong Wook Kim, Chris Hallacy, Aditya Ramesh, Gabriel Goh, Sandhini Agarwal, Girish Sastry, Amanda Askell, Pamela Mishkin, Jack Clark, et al. Learning transferable visual models from natural language supervision. In International conference on machine learning, pages 8748–8763. PMLR, 2021.
Rodrigo Santa Cruz, Léo Lebrat, Darren Fu, Pierrick Bourgeat, Jurgen Fripp, Clinton Fookes, and Olivier Salvado. Corticalflow++: Boosting cortical surface reconstruction accuracy, regularity, and interoperability. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 496–505. Springer, 2022.
Ziqiang Zhang, Sanyuan Chen, Long Zhou, Yu Wu, Shuo Ren, Shujie Liu, Zhuoyuan Yao, Xun Gong, Lirong Dai, Jinyu Li, et al. Speechlm: Enhanced speech pre-training with unpaired textual data. arXiv preprint arXiv:2209.15329, 2022.
Can Zhao, Blake E Dewey, Dzung L Pham, Peter A Calabresi, Daniel S Reich, and Jerry L Prince. Smore: a self-supervised anti-aliasing and super-resolution algorithm for mri using deep learning. IEEE transactions on medical imaging, 40(3):805–817, 2020.
Acknowledgement
This work is supported by Key-Area Research and Development Program of Guangdong Province (2023B0303040001), Guangdong Basic and Applied Basic Research Foundation (2024A1515010180) and Guangdong Provincial Key Laboratory of Human Digital Twin (2022B1212010004).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Ethics declarations
Disclosure of Interests
The authors have no competing interests to declare that are relevant to the content of this article.
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wu, W. et al. (2024). Cortical Surface Reconstruction from 2D MRI with Segmentation-Constrained Super-Resolution and Representation Learning. In: Linguraru, M.G., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2024. MICCAI 2024. Lecture Notes in Computer Science, vol 15002. Springer, Cham. https://doi.org/10.1007/978-3-031-72069-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-031-72069-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-72068-0
Online ISBN: 978-3-031-72069-7
eBook Packages: Computer ScienceComputer Science (R0)