Abstract
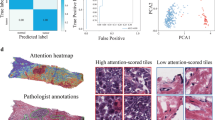
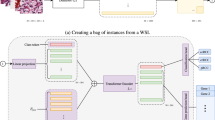
Prediction of genetic biomarkers, e.g., microsatellite instability and BRAF in colorectal cancer is crucial for clinical decision making. In this paper, we propose a whole slide image (WSI) based genetic biomarker prediction method via prompting techniques. Our work aims at addressing the following challenges: (1) extracting foreground instances related to genetic biomarkers from gigapixel WSIs, and (2) the interaction among the fine-grained pathological components in WSIs. Specifically, we leverage large language models to generate medical prompts that serve as prior knowledge in extracting instances associated with genetic biomarkers. We adopt a coarse-to-fine approach to mine biomarker information within the tumor microenvironment. This involves extracting instances related to genetic biomarkers using coarse medical prior knowledge, grouping pathology instances into fine-grained pathological components and mining their interactions. Experimental results on two colorectal cancer datasets show the superiority of our method, achieving 91.49% in AUC for MSI classification. The analysis further shows the clinical interpretability of our method. Code is publicly available at https://github.com/DeepMed-Lab-ECNU/PromptBio.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Ajani, J.A., D’Amico, T.A., Bentrem, D.J., Cooke, D., Corvera, C., Das, P., Enzinger, P.C., Enzler, T., Farjah, F., Gerdes, H., et al.: Esophageal and esophagogastric junction cancers, version 2.2023, nccn clinical practice guidelines in oncology. Journal of the National Comprehensive Cancer Network 21(4), 393–422 (2023)
Becht, E., de Reyniès, A., Giraldo, N.A., Pilati, C., Buttard, B., Lacroix, L., Selves, J., Sautès-Fridman, C., Laurent-Puig, P., Fridman, W.H.: Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clinical cancer research 22(16), 4057–4066 (2016)
Bożyk, A., Wojas-Krawczyk, K., Krawczyk, P., Milanowski, J.: Tumor microenvironment-a short review of cellular and interaction diversity. Biology 11(6), 929 (2022)
Brown, K.M., Xue, A., Smith, R.C., Samra, J.S., Gill, A.J., Hugh, T.J.: Cancer-associated stroma reveals prognostic biomarkers and novel insights into the tumour microenvironment of colorectal cancer and colorectal liver metastases. Cancer Medicine 11(2), 492–506 (2022)
Bussard, K.M., Mutkus, L., Stumpf, K., Gomez-Manzano, C., Marini, F.C.: Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Research 18, 1–11 (2016)
Caputo, F., Santini, C., Bardasi, C., Cerma, K., Casadei-Gardini, A., Spallanzani, A., Andrikou, K., Cascinu, S., Gelsomino, F.: Braf-mutated colorectal cancer: clinical and molecular insights. International journal of molecular sciences 20(21), 5369 (2019)
Chang, L., Chang, M., Chang, H.M., Chang, F.: Microsatellite instability: a predictive biomarker for cancer immunotherapy. Applied Immunohistochemistry & Molecular Morphology 26(2), e15–e21 (2018)
Dedeurwaerdere, F., Claes, K.B., Van Dorpe, J., Rottiers, I., Van der Meulen, J., Breyne, J., Swaerts, K., Martens, G.: Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Scientific reports 11(1), 12880 (2021)
Hou, W., He, Y., Yao, B., Yu, L., Yu, R., Gao, F., Wang, L.: Multi-scope analysis driven hierarchical graph transformer for whole slide image based cancer survival prediction. In: MICCAI (2023)
Huang, Z., Bianchi, F., Yuksekgonul, M., Montine, T.J., Zou, J.: A visual–language foundation model for pathology image analysis using medical twitter. Nature medicine 29(9), 2307–2316 (2023)
Ilse, M., Tomczak, J., Welling, M.: Attention-based deep multiple instance learning. ICML (2018)
Kather, J.N., Pearson, A.T., Halama, N., Jäger, D., Krause, J., Loosen, S.H., Marx, A., Boor, P., Tacke, F., Neumann, U.P., et al.: Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nature medicine 25(7), 1054–1056 (2019)
Koncina, E., Haan, S., Rauh, S., Letellier, E.: Prognostic and predictive molecular biomarkers for colorectal cancer: updates and challenges. Cancers 12(2), 319 (2020)
Li, B., Li, Y., Eliceiri, K.W.: Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. In: CVPR (2021)
Li, H., Yang, F., Zhao, Y., Xing, X., Zhang, J., Gao, M., Huang, J., Wang, L., Yao, J.: Dt-mil: deformable transformer for multi-instance learning on histopathological image. In: MICCAI. Springer (2021)
Li, H., Zhu, C., Zhang, Y., Sun, Y., Shui, Z., Kuang, W., Zheng, S., Yang, L.: Task-specific fine-tuning via variational information bottleneck for weakly-supervised pathology whole slide image classification. In: CVPR (2023)
Lin, T., Yu, Z., Hu, H., Xu, Y., Chen, C.W.: Interventional bag multi-instance learning on whole-slide pathological images. In: CVPR (2023)
Lu, M.Y., Williamson, D.F., Chen, T.Y., Chen, R.J., Barbieri, M., Mahmood, F.: Data-efficient and weakly supervised computational pathology on whole-slide images. Nature biomedical engineering 5(6), 555–570 (2021)
Macenko, M., Niethammer, M., Marron, J.S., Borland, D., Woosley, J.T., Guan, X., Schmitt, C., Thomas, N.E.: A method for normalizing histology slides for quantitative analysis. In: ISBI (2009)
Qu, L., Fu, K., Wang, M., Song, Z., et al.: The rise of ai language pathologists: Exploring two-level prompt learning for few-shot weakly-supervised whole slide image classification. Advances in Neural Information Processing Systems 36 (2024)
Qu, L., Yang, Z., Duan, M., Ma, Y., Wang, S., Wang, M., Song, Z.: Boosting whole slide image classification from the perspectives of distribution, correlation and magnification. In: ICCV (2023)
Saillard, C., Dubois, R., Tchita, O., Loiseau, N., Garcia, T., Adriansen, A., Carpentier, S., Reyre, J., Enea, D., von Loga, K., et al.: Validation of msintuit as an ai-based pre-screening tool for msi detection from colorectal cancer histology slides. Nature Communications 14(1), 6695 (2023)
Shao, Z., Bian, H., Chen, Y., Wang, Y., Zhang, J., Ji, X., et al.: Transmil: Transformer based correlated multiple instance learning for whole slide image classification. Advances in neural information processing systems 34, 2136–2147 (2021)
Shimada, Y., Okuda, S., Watanabe, Y., Tajima, Y., Nagahashi, M., Ichikawa, H., Nakano, M., Sakata, J., Takii, Y., Kawasaki, T., et al.: Histopathological characteristics and artificial intelligence for predicting tumor mutational burden-high colorectal cancer. Journal of gastroenterology 56(6), 547–559 (2021)
Sung, H., Ferlay, J., Siegel, R.L., Laversanne, M., Soerjomataram, I., Jemal, A., Bray, F.: Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 71(3), 209–249 (2021)
Tang, W., Huang, S., Zhang, X., Zhou, F., Zhang, Y., Liu, B.: Multiple instance learning framework with masked hard instance mining for whole slide image classification. In: ICCV (2023)
Tsai, P.C., Lee, T.H., Kuo, K.C., Su, F.Y., Lee, T.L.M., Marostica, E., Ugai, T., Zhao, M., Lau, M.C., Väyrynen, J.P., et al.: Histopathology images predict multi-omics aberrations and prognoses in colorectal cancer patients. Nature communications 14(1), 2102 (2023)
Zhang, H., Meng, Y., Zhao, Y., Qiao, Y., Yang, X., Coupland, S.E., Zheng, Y.: Dtfd-mil: Double-tier feature distillation multiple instance learning for histopathology whole slide image classification. In: CVPR (2022)
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 62101191), Shanghai Natural Science Foundation (Grant No. 21ZR1420800), and the Science and Technology Commission of Shanghai Municipality (Grant No. 22DZ2229004).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
Disclosure of Interests
The authors have no competing interests to declare that are relevant to the content of this article.
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, L., Yun, B., Xie, X., Li, Q., Li, X., Wang, Y. (2024). Prompting Whole Slide Image Based Genetic Biomarker Prediction. In: Linguraru, M.G., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2024. MICCAI 2024. Lecture Notes in Computer Science, vol 15004. Springer, Cham. https://doi.org/10.1007/978-3-031-72083-3_38
Download citation
DOI: https://doi.org/10.1007/978-3-031-72083-3_38
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-72082-6
Online ISBN: 978-3-031-72083-3
eBook Packages: Computer ScienceComputer Science (R0)