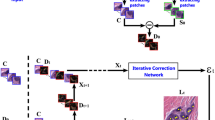
Abstract
A global threshold (e.g., 0.5) is often applied to determine which bounding boxes should be included in the final results for an object detection task. A higher threshold reduces false positives but may result in missing a significant portion of true positives. A lower threshold can increase detection recall but may also result in more false positives. Because of this, using a preset global threshold (e.g., 0.5) applied to all the bounding box candidates may lead to suboptimal solutions. In this paper, we propose a Test-time Self-guided Bounding-box Propagation (TSBP) method, leveraging Earth Mover’s Distance (EMD) to enhance object detection in histology images. TSBP utilizes bounding boxes with high confidence to influence those with low confidence, leveraging visual similarities between them. This propagation mechanism enables bounding boxes to be selected in a controllable, explainable, and robust manner, which surpasses the effectiveness of using simple thresholds and uncertainty calibration methods. Importantly, TSBP does not necessitate additional labeled samples for model training or parameter estimation, unlike calibration methods. We conduct experiments on gland detection and cell detection tasks in histology images. The results show that our proposed TSBP significantly improves detection outcomes when working in conjunction with state-of-the-art deep learning-based detection networks. Compared to other methods such as uncertainty calibration, TSBP yields more robust and accurate object detection predictions while using no additional labeled samples. The code is available at https://github.com/jwhgdeu/TSBP.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chen, S., Sun, P., Song, Y., Luo, P.: Diffusiondet: Diffusion model for object detection. In: ICCV. pp. 19773–19786 (2023)
Corbière, C., Thome, N., Bar-Hen, A., Cord, M., Pérez, P.: Addressing failure prediction by learning model confidence. Advances in Neural Information Processing Systems 32 (2019)
Davri, A., Birbas, E., Kanavos, T., Ntritsos, G., Giannakeas, N., Tzallas, A.T., Batistatou, A.: Deep learning on histopathological images for colorectal cancer diagnosis: A systematic review. Diagnostics 12(4), 837 (2022)
Echle, A., Rindtorff, N.T., Brinker, T.J., Luedde, T., Pearson, A.T., Kather, J.N.: Deep learning in cancer pathology: A new generation of clinical biomarkers. British Journal of Cancer 124(4), 686–696 (2021)
Erhan, D., Szegedy, C., Toshev, A., Anguelov, D.: Scalable object detection using deep neural networks. In: CVPR. pp. 2155–2162. IEEE Computer Society (2014)
Gilg, J., Teepe, T., Herzog, F., Rigoll, G.: The box size confidence bias harms your object detector. In: WACV. pp. 1471–1480 (2023)
Guo, C., Pleiss, G., Sun, Y., Weinberger, K.Q.: On calibration of modern neural networks. In: International Conference on Machine Learning. pp. 1321–1330. PMLR (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: CVPR. pp. 770–778 (2016)
Hendrycks, D., Gimpel, K.: A baseline for detecting misclassified and out-of-distribution examples in neural networks. In: ICLR (2017)
Kull, M., Silva Filho, T., Flach, P.: Beta calibration: A well-founded and easily implemented improvement on logistic calibration for binary classifiers. In: Artificial Intelligence and Statistics. pp. 623–631. PMLR (2017)
Kumar, N., Verma, R., Sharma, S., Bhargava, S., Vahadane, A., Sethi, A.: A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Transactions on Medical Imaging 36(7), 1550–1560 (2017)
Kuppers, F., Kronenberger, J., Shantia, A., Haselhoff, A.: Multivariate confidence calibration for object detection. In: CVPR Workshops. pp. 326–327 (2020)
Van der Laak, J., Litjens, G., Ciompi, F.: Deep learning in histopathology: The path to the clinic. Nature Medicine 27(5), 775–784 (2021)
Munir, M.A., Khan, M.H., Sarfraz, M.S., Ali, M.: Towards improving calibration in object detection under domain shift. In: NeurIPS (2022)
Naeini, M.P., Cooper, G., Hauskrecht, M.: Obtaining well calibrated probabilities using Bayesian binning. In: Proceedings of the AAAI Conference on Artificial Intelligence. vol. 29 (2015)
Nguyen, A., Yosinski, J., Clune, J.: Deep neural networks are easily fooled: High confidence predictions for unrecognizable images. In: CVPR. pp. 427–436 (2015)
Platt, J., et al.: Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Advances in large margin classifiers 10(3), 61–74 (1999)
Sirinukunwattana, K., Pluim, J.P., Chen, H., Qi, X., Heng, P.A., Guo, Y.B., Wang, L.Y., Matuszewski, B.J., Bruni, E., Sanchez, U., et al.: Gland segmentation in colon histology images: The GlaS challenge contest. Medical Image Analysis 35, 489–502 (2017)
Wenkel, S., Alhazmi, K., Liiv, T., Alrshoud, S., Simon, M.: Confidence score: The forgotten dimension of object detection performance evaluation. Sensors 21(13), 4350 (2021)
Yang, X., Wu, J., He, L., Ma, S., Hou, Z., Sun, W.: CPSS-FAT: A consistent positive sample selection for object detection with full adaptive threshold. Pattern Recognition 141, 109627 (2023)
Yue, X., Li, H., Shimizu, M., Kawamura, S., Meng, L.: Deep learning-based real-time object detection for empty-dish recycling robot. In: 2022 13th Asian Control Conference (ASCC). pp. 2177–2182. IEEE (2022)
Zadrozny, B., Elkan, C.: Obtaining calibrated probability estimates from decision trees and naive bayesian classifiers. In: ICML. vol. 1, pp. 609–616 (2001)
Acknowledgments
We sincerely thank the reviewers for their time and effort in reviewing our manuscript and for providing constructive feedback to improve our work. This research was supported in part by the Natural Science Foundation of Jiangsu Province (Grant BK20220949), and National Natural Science Foundation of China (Grant 62201263).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Ethics declarations
Disclosure of Interests
The authors have no competing interests to declare.
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Yang, T., Xiao, L., Zhang, Y. (2024). TSBP: Improving Object Detection in Histology Images via Test-Time Self-guided Bounding-Box Propagation. In: Linguraru, M.G., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2024. MICCAI 2024. Lecture Notes in Computer Science, vol 15004. Springer, Cham. https://doi.org/10.1007/978-3-031-72083-3_49
Download citation
DOI: https://doi.org/10.1007/978-3-031-72083-3_49
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-72082-6
Online ISBN: 978-3-031-72083-3
eBook Packages: Computer ScienceComputer Science (R0)