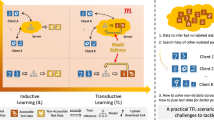
Abstract
Federated learning is one popular paradigm to train a joint model in a distributed, privacy-preserving environment. But partial annotations pose an obstacle meaning that categories of labels are heterogeneous over clients. We propose to learn a joint backbone in a federated manner, while each site receives its own multi-label segmentation head. By using Bayesian techniques we observe that the different segmentation heads although only trained on the individual client’s labels also learn information about the other labels not present at the respective site. This information is encoded in their predictive uncertainty. To obtain a final prediction we leverage this uncertainty and perform a weighted averaging of the ensemble of distributed segmentation heads, which allows us to segment “locally unknown” structures. With our method, which we refer to as FUNAvg, we are even on-par with the models trained and tested on the same dataset on average. The code is publicly available (https://github.com/Cardio-AI/FUNAvg).
B. Menze and S. Engelhardt—These authors contributed equally.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bilic, P., Christ, P., Li, H.B., Menze, B., et al.: The liver tumor segmentation benchmark (LiTS). Med. Image Anal. 84, 102680 (2023). https://doi.org/10.1016/j.media.2022.102680
Boughorbel, S., Jarray, F., Venugopal, N., Moosa, S., Elhadi, H., Makhlouf, M.: Federated uncertainty-aware learning for distributed hospital EHR data. In: Machine Learning for Health (ML4H) at NeurIPS (2019)
Chen, H.Y., Chao, W.L.: FedBE: making Bayesian model ensemble applicable to federated learning. In: ICLR (2021). https://doi.org/10.48550/arXiv.2009.01974
Fuchs, M., González, C., Mukhopadhyay, A.: Practical uncertainty quantification for brain tumor segmentation. In: MIDL, vol. 172, pp. 407–422 (2022)
Gal, Y., Ghahramani, Z.: Dropout as a Bayesian approximation: representing model uncertainty in deep learning. In: ICML, vol. 48, pp. 1050–1059 (2016). https://doi.org/10.5555/3045390.3045502
Guo, C., Pleiss, G., Sun, Y., Weinberger, K.Q.: On calibration of modern neural networks. In: ICML, vol. 70, pp. 1321–1330 (2017). https://doi.org/10.5555/3305381.3305518
Hatamizadeh, A., Nath, V., Tang, Y., Yang, D., Roth, H.R., Xu, D.: Swin UNETR: Swin transformers for semantic segmentation of brain tumors in MRI images. In: Crimi, A., Bakas, S. (eds.) BrainLes 2021. LNCS, vol. 12962, pp. 272–284. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-08999-2_22
Isensee, F., Jaeger, P.F., Kohl, S.A.A., Petersen, J., Maier-Hein, K.H.: nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021). https://doi.org/10.1038/s41592-020-01008-z
Ji, Y., et al.: AMOS: a large-scale abdominal multi-organ benchmark for versatile medical image segmentation. In: NeuRIPS, vol. 35, pp. 36722–36732 (2022). https://doi.org/10.5281/zenodo.7262581
Karimireddy, S.P., Kale, S., Mohri, M., Reddi, S., Stich, S., Suresh, A.T.: SCAFFOLD: stochastic controlled averaging for federated learning. In: ICML, vol. 119, pp. 5132–5143 (2020)
Kavur, A.E., et al.: CHAOS challenge - combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 69, 101950 (2021). https://doi.org/10.1016/j.media.2020.101950
Kendall, A., Gal, Y.: What uncertainties do we need in Bayesian deep learning for computer vision? In: NeuRIPS, vol. 30 (2017)
Kohl, S., et al.: A probabilistic U-Net for segmentation of ambiguous images. In: NeuRIPS, vol. 31 (2018)
Kwon, Y., Won, J.H., Kim, B.J., Paik, M.C.: Uncertainty quantification using Bayesian neural networks in classification: application to ischemic stroke lesion segmentation. In: MIDL (2018). https://openreview.net/forum?id=Sk_P2Q9sG
Landman, B., Xu, Z., Igelsias, J., Styner, M., Langerak, T., Klein, A.: MICCAI multi-atlas labeling beyond the cranial vault-workshop and challenge. In: MICCAI Multi-Atlas Labeling Beyond Cranial Vault-Workshop Challenge (2015). https://doi.org/10.7303/SYN3193805
Laves, M.H., Tölle, M., Schlaefer, A., Engelhardt, S.: Posterior temperature optimized Bayesian models for inverse problems in medical imaging. Med. Image Anal. 78, 102382 (2022). https://doi.org/10.1016/j.media.2022.102382
Li, T., Sahu, A.K., Zaheer, M., Sanjabi, M., Talwalkar, A., Smith, V.: Federated optimization in heterogeneous networks. In: MLSys (2020)
Linsner, F., Adilova, L., Däubener, S., Kamp, M., Fischer, A.: Approaches to uncertainty quantification in federated deep learning. In: Kamp, M., et al. (eds.) ECML PKDD 2021. LNCS, vol. 1524, pp. 128–145. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-93736-2_12
Ma, J., et al.: AbdomenCT-1K: is abdominal organ segmentation a solved problem? IEEE Trans. Pattern Anal. Mach. Intell. 44(10), 6695–6714 (2022). https://doi.org/10.1109/TPAMI.2021.3100536
McMahan, H.B., Moore, E., Ramage, D., Hampson, S., y Arcas, B.A.: Communication-efficient learning of deep networks from decentralized data. In: AISTATS (2017)
Oktay, O., et al.: Anatomically constrained neural networks (ACNN): application to cardiac image enhancement and segmentation. IEEE Trans. Med. Imaging 37(2), 384–395 (2018). https://doi.org/10.1109/TMI.2017.2743464
Rädsch, T., et al.: Labelling instructions matter in biomedical image analysis. Nat. Mach. Intell. 5(3), 273–283 (2023). https://doi.org/10.1038/s42256-023-00625-5
Thorgeirsson, A.T., Gauterin, F.: Probabilistic predictions with federated learning. Entropy 23(1), 41 (2020). https://doi.org/10.3390/e23010041
Ulrich, C., Isensee, F., Wald, T., Zenk, M., Baumgartner, M., Maier-Hein, K.H.: MultiTalent: a multi-dataset approach to medical image segmentation. In: Greenspan, H., et al. (eds.) MICCAI 2023. LNCS, vol. 14222, pp. 648–658. Springer, Cham (2023). https://doi.org/10.1007/978-3-031-43898-1_62
Wasserthal, J., et al.: TotalSegmentator: robust segmentation of 104 anatomical structures in CT images. Radiol. Artif. Intell. 5(5) (2023). https://doi.org/10.1148/ryai.230024
Xu, X., Deng, H.H., Gateno, J., Yan, P.: Federated multi-organ segmentation with inconsistent labels. Trans. Med. Imaging 42(10), 2948–2960 (2023). https://doi.org/10.1109/TMI.2023.3270140
Xu, Z., et al.: Evaluation of six registration methods for the human abdomen on clinically acquired CT. Trans. Biomed. Eng. 63(8), 1563–1572 (2016). https://doi.org/10.1109/TBME.2016.2574816
Zhou, Y., et al.: Prior-aware neural network for partially-supervised multi-organ segmentation. In: ICCV (2019). https://doi.org/10.1109/ICCV.2019.01077
Acknowledgments
The work was done during a research stay of MT at Menze lab at University Zurich (UZH) with support from the DAAD (German Academic Exchange Service). MT and SE are supported by grants from the Klaus Tschira Foundation within the Informatics for Life framework, by the DZHK and BMBF, in particular BMBF-SWAG Project 01KD2215D. FN and BM are supported by the Helmut Horten foundation. The authors gratefully acknowledge the data storage service SDS@hd supported by MWK and DFG through grant INST 35/1314-1 FUGG and INST 35/1503-1 FUGG.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
Disclosure of Interests
The authors have no competing interests to declare that are relevant to the content of this article.
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Tölle, M., Navarro, F., Eble, S., Wolf, I., Menze, B., Engelhardt, S. (2024). FUNAvg: Federated Uncertainty Weighted Averaging for Datasets with Diverse Labels. In: Linguraru, M.G., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2024. MICCAI 2024. Lecture Notes in Computer Science, vol 15010. Springer, Cham. https://doi.org/10.1007/978-3-031-72117-5_38
Download citation
DOI: https://doi.org/10.1007/978-3-031-72117-5_38
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-72116-8
Online ISBN: 978-3-031-72117-5
eBook Packages: Computer ScienceComputer Science (R0)