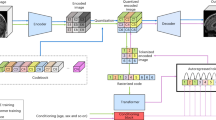
Abstract
Synthetic longitudinal brain MRI simulates brain aging and would enable more efficient research on neurodevelopmental and neurodegenerative conditions. Synthetically generated, age-adjusted brain images could serve as valuable alternatives to costly longitudinal imaging acquisitions, serve as internal controls for studies looking at the effects of environmental or therapeutic modifiers on brain development, and allow data augmentation for diverse populations. In this paper, we present a diffusion-based approach called SynthBrainGrow for synthetic brain aging with a two-year step. To validate the feasibility of using synthetically generated data on downstream tasks, we compared structural volumetrics of two-year-aged brains against synthetically aged brain MRI. The use of structural similarity indices, such as the Structural Similarity Index Measure (SSIM), for evaluating synthetic medical images has come under recent scrutiny. These indices may not effectively capture the perceptual quality or clinical usefulness in synthesized radiology scans. To assess the performance of SynthBrainGrow, we evaluated the substructural volumetric similarity between synthetic and real patient scans. Results show that SynthBrainGrow can accurately capture substructure volumetrics and simulate structural changes such as ventricle enlargement and cortical thinning. Generating longitudinal brain datasets from cross-sectional data could enable augmented training and benchmarking of computational tools for analyzing lifespan trajectories. This work signifies an important advance in generative modeling to synthesize realistic longitudinal data with limited lifelong MRI scans. The code is available at https://github.com/zapaishchykova/SynthBrainGrow.
A. Zapaishchykova and B. H. Kann—These authors contributed equally to this manuscript.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
R. a. I. Bethlehem et al., “Brain charts for the human lifespan,” Nature, vol. 604, no. 7906, Art. no. 7906, Apr. 2022, https://doi.org/10.1038/s41586-022-04554-y.
J. H. Cole et al., “Brain age predicts mortality,” Mol. Psychiatry, vol. 23, no. 5, Art. no. 5, May 2018, https://doi.org/10.1038/mp.2017.62.
E. L. Grigorenko, “Brain Development: The Effect of Interventions on Children and Adolescents,” in Child and Adolescent Health and Development, 3rd ed., D. A. P. Bundy, N. de Silva, S. Horton, D. T. Jamison, and G. C. Patton, Eds., Washington (DC): The International Bank for Reconstruction and Development / The World Bank, 2017. Accessed: Jan. 31, 2024. [Online]. Available: http://www.ncbi.nlm.nih.gov/books/NBK525261/
W. H. L. Pinaya et al., “Brain Imaging Generation with Latent Diffusion Models,” in Deep Generative Models, A. Mukhopadhyay, I. Oksuz, S. Engelhardt, D. Zhu, and Y. Yuan, Eds., in Lecture Notes in Computer Science. Cham: Springer Nature Switzerland, 2022, pp. 117–126. https://doi.org/10.1007/978-3-031-18576-2_12.
B. J. Casey et al., “The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites,” Dev. Cogn. Neurosci., vol. 32, pp. 43–54, Aug. 2018, https://doi.org/10.1016/j.dcn.2018.03.001.
L. M. Wierenga et al., “Unraveling age, puberty and testosterone effects on subcortical brain development across adolescence,” Psychoneuroendocrinology, vol. 91, pp. 105–114, May 2018, https://doi.org/10.1016/j.psyneuen.2018.02.034.
Y. Zhou, N. Pat, and M. C. Neale, “Associations between resting state functional brain connectivity and childhood anhedonia: A reproduction and replication study,” PLOS ONE, vol. 18, no. 5, p. e0277158, May 2023, https://doi.org/10.1371/journal.pone.0277158.
P. Dhariwal and A. Nichol, “Diffusion Models Beat GANs on Image Synthesis.” arXiv, Jun. 01, 2021. https://doi.org/10.48550/arXiv.2105.05233.
A. Q. Nichol and P. Dhariwal, “Improved Denoising Diffusion Probabilistic Models,” in Proceedings of the 38th International Conference on Machine Learning, PMLR, Jul. 2021, pp. 8162–8171. Accessed: Jan. 10, 2024. [Online]. Available: https://proceedings.mlr.press/v139/nichol21a.html
J. Wolleb, R. Sandkühler, F. Bieder, P. Valmaggia, and P. C. Cattin, “Diffusion Models for Implicit Image Segmentation Ensembles.” arXiv, Dec. 27, 2021. https://doi.org/10.48550/arXiv.2112.03145.
Y. Xie and Q. Li, “Measurement-conditioned Denoising Diffusion Probabilistic Model for Under-sampled Medical Image Reconstruction.” arXiv, Mar. 05, 2022. https://doi.org/10.48550/arXiv.2203.03623.
A. Zapaishchykova et al., “Diffusion Deep Learning for Brain Age Prediction and Longitudinal Tracking in Children Through Adulthood.” medRxiv, p. 2023.10.17.23297166, Oct. 20, 2023. https://doi.org/10.1101/2023.10.17.23297166.
A. Durrer et al., “Diffusion Models for Contrast Harmonization of Magnetic Resonance Images.” arXiv, Mar. 14, 2023. https://doi.org/10.48550/arXiv.2303.08189.
S. Bao et al., “Prediction of brain age using quantitative parameters of synthetic magnetic resonance imaging,” Front. Aging Neurosci., vol. 14, 2022, Accessed: Jan. 12, 2024. [Online]. Available: https://www.frontiersin.org/articles/https://doi.org/10.3389/fnagi.2022.963668
J. Fu et al., “Fast three-dimensional image generation for healthy brain aging using diffeomorphic registration,” Hum. Brain Mapp., vol. 44, no. 4, pp. 1289–1308, 2023, https://doi.org/10.1002/hbm.26165.
G. Pombo et al., “Equitable modelling of brain imaging by counterfactual augmentation with morphologically constrained 3D deep generative models,” Med. Image Anal., vol. 84, p. 102723, Feb. 2023, https://doi.org/10.1016/j.media.2022.102723.
J. Wang, M. N. Lytle, Y. Weiss, B. L. Yamasaki, and J. R. Booth, “A longitudinal neuroimaging dataset on language processing in children ages 5, 7, and 9 years old,” Sci. Data, vol. 9, no. 1, Art. no. 1, Jan. 2022, https://doi.org/10.1038/s41597-021-01106-3.
Z. Dorjsembe, H.-K. Pao, S. Odonchimed, and F. Xiao, “Conditional Diffusion Models for Semantic 3D Medical Image Synthesis.” arXiv, Jul. 31, 2023. https://doi.org/10.48550/arXiv.2305.18453.
S. Klein, M. Staring, K. Murphy, M. A. Viergever, and J. P. W. Pluim, “elastix: A Toolbox for Intensity-Based Medical Image Registration,” IEEE Trans. Med. Imaging, vol. 29, no. 1, pp. 196–205, Jan. 2010, https://doi.org/10.1109/TMI.2009.2035616.
V. Fonov, A. C. Evans, K. Botteron, C. R. Almli, R. C. McKinstry, and D. L. Collins, “Unbiased average age-appropriate atlases for pediatric studies,” NeuroImage, vol. 54, no. 1, pp. 313–327, Jan. 2011, https://doi.org/10.1016/j.neuroimage.2010.07.033.
F. Isensee et al., “Automated brain extraction of multisequence MRI using artificial neural networks,” Hum. Brain Mapp., vol. 40, no. 17, pp. 4952–4964, 2019, https://doi.org/10.1002/hbm.24750.
W. H. L. Pinaya et al., “Generative AI for Medical Imaging: extending the MONAI Framework.” arXiv, Jul. 27, 2023. https://doi.org/10.48550/arXiv.2307.15208.
“SynthSR: A public AI tool to turn heterogeneous clinical brain scans into high-resolution T1-weighted images for 3D morphometry | Science Advances.” Accessed: Jan. 12, 2024. [Online]. Available: https://www.science.org/doi/https://doi.org/10.1126/sciadv.add3607
B. Billot et al., “SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining,” Med. Image Anal., vol. 86, p. 102789, May 2023, https://doi.org/10.1016/j.media.2023.102789.
“Properties of the SSIM metric in medical image assessment: Correspondence between measurements and the spatial frequency spectrum.” Accessed: Jan. 11, 2024. [Online]. Available: https://www.researchsquare.com
“On the proper use of structural similarity for the robust evaluation of medical image synthesis models - Gourdeau - 2022 - Medical Physics - Wiley Online Library.” Accessed: Jan. 10, 2024. [Online]. Available: https://aapm.onlinelibrary.wiley.com/doi/https://doi.org/10.1002/mp.15514
C.-Y. Yang, C. Ma, and M.-H. Yang, “Single-Image Super-Resolution: A Benchmark,” in Computer Vision – ECCV 2014, D. Fleet, T. Pajdla, B. Schiele, and T. Tuytelaars, Eds., in Lecture Notes in Computer Science. Cham: Springer International Publishing, 2014, pp. 372–386. https://doi.org/10.1007/978-3-319-10593-2_25.
G. P. Renieblas, A. T. Nogués, A. M. González, N. Gómez-Leon, and E. G. Del Castillo, “Structural similarity index family for image quality assessment in radiological images,” J. Med. Imaging Bellingham Wash, vol. 4, no. 3, p. 035501, Jul. 2017, https://doi.org/10.1117/1.JMI.4.3.035501.
Acknowledgments
Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive DevelopmentSM (ABCD) Study (https://abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit more than 10,000 children age 9–10 and follow them over 10 years into early adulthood. The ABCD Study® is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, U24DA041147. A full list of supporters is available at https://abcdstudy.org/federal-partners.html. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/consortium_members/. ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in the analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Ethics declarations
Scientific Advisory Board - Day One Biopharmaceuticals; Funding: NIH/NCI U54 CA274516, P50 CA165962; Botha-Chan Low Grade Glioma Consortium.
Appendix
Appendix
Two example subjects randomly drawn from Long579 dataset. Top row: raw segmentation(using SynthSeg) on image with 3 × 3 × 2.5 mm3 in voxel size with “under-segmented” artifacts. Bottom row: we upsampled the image × 2 using spline interpolation, resampled voxel size back to 1 × 1 × 1 mm3, and increased image resolution using FreeSurfer v.7.4.1 SynthSR v2.0 [23].
The median percentage differences between predicted and ground truth(GT) values for various brain regions, along with their 95% confidence intervals(ABCD and Long579 datasets, N = 1473). The segmentation volumetrics were calculated using SynthSeg. The brain regions are categorized into four groups. Gray Matter Volume (GMV): Includes the left and right cerebral cortex. White Matter Volume (WMV): Includes the left and right cerebral white matter. Subcortical Gray Matter Volume (sGMV): Comprises regions such as the thalamus, caudate, putamen, pallidum, hippocampus, amygdala, accumbens area, and ventral DC. These regions often have less distinct boundaries than cortical or white matter regions. Many subcortical structures are smaller in volume compared to cortical and white matter regions. Even small absolute errors in segmentation can result in large percentage differences. Ventricular Volume (VV): Includes the left and right lateral ventricles and the inferior lateral ventricles.
Rights and permissions
Copyright information
© 2025 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Zapaishchykova, A., Kann, B.H., Tak, D., Ye, Z., Haas-Kogan, D.A., Aerts, H.J.W.L. (2025). SynthBrainGrow: Synthetic Diffusion Brain Aging for Longitudinal MRI Data Generation in Young People. In: Mukhopadhyay, A., Oksuz, I., Engelhardt, S., Mehrof, D., Yuan, Y. (eds) Deep Generative Models. DGM4MICCAI 2024. Lecture Notes in Computer Science, vol 15224. Springer, Cham. https://doi.org/10.1007/978-3-031-72744-3_8
Download citation
DOI: https://doi.org/10.1007/978-3-031-72744-3_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-72743-6
Online ISBN: 978-3-031-72744-3
eBook Packages: Computer ScienceComputer Science (R0)