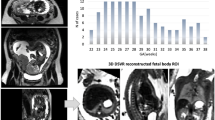
Abstract
Fetal body parts segmentation can be useful to detect abnormalities and assess fetal growth from magnetic resonance imaging (MRI). In this work, 3D fetal head, trunk and limbs body parts segmentation is leveraged for the first time by volumetric reconstructions of the whole fetal anatomy coupled to deep learning techniques for volumetric segmentation. Due to the time consuming manual segmentation required for training in this volumetric multi-label setting, sparse annotations are used, marking slices with a \(6\,\text {mm}\) separation and alternating the slicing axis between cases. These manual segmentations are used to train and compare different models using both the original MRI data and the data after volumetric reconstruction in a dataset of 45 cases. The setup consisting on 3D volumetric reconstructions and a 3D U-net based learning model results in optimal segmentation metrics, with Dice scores higher than 0.9 for all the considered structures and 0.973 for the fetal body (0.979 when highly motion corrupted datasets are discarded). In comparison, the performance of the setups that use the original MRI data exhibit a pronounced decline in segmentation scores, highlighting the importance of robust reconstruction techniques for automatic fetal growth characterization. Finally, we conduct a Bland-Altman analysis studying the reliability of our proposed 3D reconstruction and segmentation pipeline for automatic estimation of fetal body part weights.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Papaioannou, G., Caro-Domínguez, P., Klein, W.M., Garel, C., Cassart, M.: Indications for magnetic resonance imaging of the fetal body (extra-central nervous system): recommendations from the European Society of Paediatric Radiology Fetal Task Force. Pediatr. Radiol. 53(2), 297–312 (2023). https://doi.org/10.1007/s00247-022-05495-4
Wilson, L., Whitby, E.H.: The value of fetal magnetic resonance imaging in diagnosis of congenital anomalies of the fetal body: a systematic review and meta-analysis. BMC Med. Imag. 24(1) (2024). https://doi.org/10.1186/s12880-024-01286-5
Uus, A., et al.: Deformable slice-to-volume registration for motion correction of fetal body and placenta MRI. IEEE Trans. Med. Imaging 39(9), 2750–2759 (2020). https://doi.org/10.1109/tmi.2020.2974844
Mufti, N., et al.: Use of super resolution reconstruction MRI for surgical planning in Placenta accreta spectrum disorder: case series. Placenta 142, 36–45 (2023). https://doi.org/10.1016/j.placenta.2023.08.066
Cordero-Grande, L., et al.: Fetal MRI by robust deep generative prior reconstruction and diffeomorphic registration. IEEE Trans. Med. Imaging 42(3), 810–822 (2023). https://doi.org/10.1109/tmi.2022.3217725
Uus, A.U., et al.: Automated body organ segmentation, volumetry and population-averaged atlas for 3D motion-corrected T2-weighted fetal body MRI. Sci. Rep. 14(1) (2024). https://doi.org/10.1038/s41598-024-57087-x
Ryd, D., Nilsson, A., Heiberg, E., Hedström, E.: Automatic segmentation of the fetus in 3D magnetic resonance images using deep learning: accurate and fast fetal volume quantification for clinical use. Pediatr. Cardiol. 44(6), 1311–1318 (2023). https://doi.org/10.1007/s00246-022-03038-0
Specktor-Fadida, B., et al.: Deep learning-based segmentation of whole-body fetal MRI and fetal weight estimation: assessing performance, repeatability, and reproducibility. Eur. Radiol. 34(3), 2072–2083 (2024). https://doi.org/10.1007/s00330-023-10038-y
Rabinowich, A., et al.: Fetal MRI-based body and adiposity quantification for small for gestational age perinatal risk stratification. J. Magn. Reson. Imaging 60(2), 767–774 (2024). https://doi.org/10.1002/jmri.29141
Hall, M., et al.: Adrenal volumes in fetuses delivering prior to 32.weeks’ gestation: an MRI pilot study. Acta Obstetricia et Gynecologica Scandinavica 103(3), 512–521 (2024). https://doi.org/10.1111/aogs.14733
Zhang, T., et al.: Graph-based whole body segmentation in fetal MR images. In: MICCAI Workshop PIPPI (2016). https://pippiworkshop.github.io/pippi2016/pdf/PIPPI2016_04_Zhang.pdf
Kulseng, C.P.S., Hillestad, V., Eskild, A., Gjesdal, K.I.: Automatic placental and fetal volume estimation by a convolutional neural network. Placenta 134, 23–29 (2023). https://doi.org/10.1016/j.placenta.2023.02.009
Dudovitch, G., Link-Sourani, D., Ben Sira, L., Miller, E., Ben Bashat, D., Joskowicz, L.: Deep learning automatic fetal structures segmentation in MRI scans with few annotated datasets. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12266, pp. 365–374. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59725-2_35
Lo, J., et al.: Cross attention squeeze excitation network (CASE-Net) for whole body fetal MRI segmentation. Sensors 21(13) (2021). https://doi.org/10.3390/s21134490
Fadida, B.S., Sourani, D.L., Sira, L.B., Miller, E., Bashat, D.B., Joskowicz, L.: Partial annotations for the segmentation of large structures with low annotation cost. In: Zamzmi, G., Antani, S., Bagci, U., Linguraru, M.G., Rajaraman, S., Xue, Z. (eds.) MILLanD 2022. LNCS, vol. 13559, pp. 13–22. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-16760-7_2
iFIND: Intelligent fetal imaging and diagnosis. http://www.ifindproject.com
Isensee, F., Jaeger, P.F., Kohl, S.A.A., Petersen, J., Maier-Hein, K.H.: nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021). https://doi.org/10.1038/s41592-020-01008-z
Isensee, F., et al.: nnU-Net revisited: a call for rigorous validation in 3D medical image segmentation (2024). https://doi.org/10.48550/arXiv.2404.09556
Baker, P., et al.: Fetal weight estimation by echo-planar magnetic resonance imaging. Lancet 343(8898), 644–645 (1994). https://doi.org/10.1016/s0140-6736(94)92638-7
Liu, R., et al.: An intriguing failing of convolutional neural networks and the CoordConv solution, July 2018. https://doi.org/10.48550/arXiv.1807.03247
Acknowledgments
This work was supported in part by MCIN, Spain, under the Beatriz Galindo Program (BGP18/00178) and the STEP-AMD project (TED2021-1319518-I00); in part by MCIN/AEI/10.13039/5011000110 33/FEDER, EU, under Projects PID2021-129022OA-I00 and PID2022-141493OB-I00; and in part by the Madrid Government, Spain, under the Multiannual Agreement with Universidad Politécnica de Madrid in the line support for Research and Development Projects for Beatriz Galindo Researchers, in the context of V PRICIT. The authors gratefully acknowledge the Universidad Politécnica de Madrid for providing computing resources on Magerit Supercomputer.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
Disclosure of Interests
The authors have no competing interests to declare that are relevant to the content of this article.
Rights and permissions
Copyright information
© 2025 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Alarcón-Gil, P.P., Alfano, F., Uus, A., Ledesma-Carbayo, M.J., Cordero-Grande, L. (2025). Fetal Body Parts Segmentation Using Volumetric MRI Reconstructions. In: Link-Sourani, D., Abaci Turk, E., Macgowan, C., Hutter, J., Melbourne, A., Licandro, R. (eds) Perinatal, Preterm and Paediatric Image Analysis. PIPPI 2024. Lecture Notes in Computer Science, vol 14747. Springer, Cham. https://doi.org/10.1007/978-3-031-73260-7_12
Download citation
DOI: https://doi.org/10.1007/978-3-031-73260-7_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-73259-1
Online ISBN: 978-3-031-73260-7
eBook Packages: Computer ScienceComputer Science (R0)