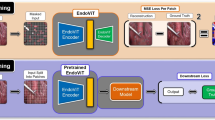
Abstract
Over the past decade, computer vision applications in minimally invasive surgery have rapidly increased. Despite this growth, the impact of surgical computer vision remains limited compared to other medical fields like pathology and radiology, primarily due to the scarcity of representative annotated data. Whereas transfer learning from large annotated datasets such as ImageNet has been conventionally the norm to achieve high-performing models, recent advancements in self-supervised learning (SSL) have demonstrated superior performance. In medical image analysis, in-domain SSL pretraining has already been shown to outperform ImageNet-based initialization. Although unlabeled data in the field of surgical computer vision is abundant, the diversity within this data is limited. This study investigates the role of dataset diversity in SSL for surgical computer vision, comparing procedure-specific datasets against a more heterogeneous general surgical dataset across three different downstream surgical applications. The obtained results show that using solely procedure-specific data can lead to substantial improvements of 13.8%, 9.5%, and 36.8% compared to ImageNet pretraining. However, extending this data with more heterogeneous surgical data further increases performance by an additional 5.0%, 5.2%, and 2.5%, suggesting that increasing diversity within SSL data is beneficial for model performance. The code and pretrained model weights are made publicly available at https://github.com/TimJaspers0801/SurgeNet.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Alapatt, D., Murali, A., Srivastav, V., Mascagni, P., Consortium, A., Padoy, N.: Jumpstarting surgical computer vision (2023)
Bakker, F.H.A., de Nijs, J.V., Jaspers, T., et al.: Estimating surgical urethral length on intraoperative robot-assisted prostatectomy images using artificial intelligence anatomy recognition. J. Endourol. 38(7), 690–696 (2024). https://doi.org/10.1089/end.2023.0697, pMID: 38613819
Bawa, V.S., Singh, G., KapingA, F., et al.: The SARAS Endoscopic Surgeon Action Detection (ESAD) dataset: challenges and methods (2021)
den Boer, R.B., Jaspers, T.J.M., de Jongh, C., et al.: Deep learning-based recognition of key anatomical structures during robot-assisted minimally invasive esophagectomy. Surg. Endosc. 37(7), 5164–5175 (2023). https://doi.org/10.1007/s00464-023-09990-z
den Boer, R.B., de Jongh, C., Huijbers, W.T.E., et al.: Computer-aided anatomy recognition in intrathoracic and -abdominal surgery: a systematic review. Surg. Endosc. 36(12), 8737–8752 (2022). https://doi.org/10.1007/s00464-022-09421-5
Caron, M., Touvron, H., et al.: Emerging properties in self-supervised vision transformers. In: Proceedings of the International Conference on Computer Vision (ICCV) (2021)
Carstens, M., Rinner, F.M., Bodenstedt, S., et al.: The Dresden surgical anatomy dataset for abdominal organ segmentation in surgical data science. Sci. Data 10(1), 3 (2023). https://doi.org/10.1038/s41597-022-01719-2
Deng, J., Dong, W., Socher, R., et al.: ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255 (2009). https://doi.org/10.1109/CVPR.2009.5206848
Dosovitskiy, A., et al.: An image is worth 16 \(\times \) 16 words: transformers for image recognition at scale (2021)
Hashimoto, D.A., Rosman, G., Volkov, M., Rus, D.L., Meireles, O.R.: Artificial intelligence for intraoperative video analysis: machine learning’s role in surgical education. J. Am. Coll. Surg. 225(4, Suppl. 1), S171 (2017). https://doi.org/10.1016/j.jamcollsurg.2017.07.387, Scientific Forum Abstracts: 2017 Clinical Congress
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition (2015)
Hirsch, R., Caron, M., Cohen, R., et al.: Self-supervised learning for endoscopic video analysis. In: Greenspan, H., et al. (eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2023, pp. 569–578. Springer, Cham (2023). https://doi.org/10.1007/978-3-031-43904-9_55
Hong, W.Y., Kao, C.L., Kuo, Y.H., et al.: CholecSeg8k: a semantic segmentation dataset for laparoscopic cholecystectomy based on Cholec80 (2020)
Kirillov, A., Girshick, R., He, K., Dollar, P.: Panoptic feature pyramid networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), June 2019
Lavanchy, J.L., Ramesh, S., Dall’Alba, D., et al.: Challenges in multi-centric generalization: phase and step recognition in Roux-en-Y gastric bypass surgery. Int. J. Comput. Assist. Radiol. Surg. (2024). https://doi.org/10.1007/s11548-024-03166-3
Leibetseder, A., Kletz, S., Schoeffmann, K., Keckstein, S., Keckstein, J.: GLENDA: gynecologic laparoscopy endometriosis dataset. In: Ro, Y.M., et al. (eds.) MultiMedia Modeling, pp. 439–450. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-37734-2_36
Leibetseder, A., Petscharnig, S., Primus, M.J., et al.: LapGyn4: a dataset for 4 automatic content analysis problems in the domain of laparoscopic gynecology. In: Proceedings of the 9th ACM Multimedia Systems Conference, pp. 357–362 (2018)
Maier-Hein, L., Eisenmann, M., Sarikaya, D., et al.: Surgical data science - from concepts toward clinical translation. Med. Image Anal. 76, 102306 (2022). https://doi.org/10.1016/j.media.2021.102306
Maier-Hein, L., Wagner, M., Ross, T., et al.: Heidelberg colorectal data set for surgical data science in the sensor operating room (2021)
Mascagni, P., Vardazaryan, A., Alapatt, D., et al.: Artificial intelligence for surgical safety: automatic assessment of the critical view of safety in laparoscopic cholecystectomy using deep learning. Ann. Surg. 275(5), 955–961 (2022)
Padoy, N., Blum, T., Ahmadi, S.A., Feussner, H., Berger, M.O., Navab, N.: Statistical modeling and recognition of surgical workflow. Med. Image Anal. 16(3), 632–641 (2012). https://doi.org/10.1016/j.media.2010.10.001, Computer Assisted Interventions
Ramesh, S., Srivastav, V., Alapatt, D., et al.: Dissecting self-supervised learning methods for surgical computer vision. Med. Image Anal. 88, 102844 (2023). https://doi.org/10.1016/j.media.2023.102844
Tan, M., Le, Q.V.: EfficientNet: rethinking model scaling for convolutional neural networks (2020)
Twinanda, A.P., Shehata, S., Mutter, D., Marescaux, J., de Mathelin, M., Padoy, N.: EndoNet: a deep architecture for recognition tasks on laparoscopic videos. IEEE Trans. Med. Imaging 36(1), 86–97 (2017). https://doi.org/10.1109/TMI.2016.2593957
Valderrama, N., Ruiz Puentes, P., Hernández, I., et al.: Towards holistic surgical scene understanding. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022, pp. 442–452. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-16449-1_42
Wang, W., et al.: Pyramid vision transformer: a versatile backbone for dense prediction without convolutions (2021)
Wang, Z., Liu, C., et al.: Foundation model for endoscopy video analysis via large-scale self-supervised pre-train. In: Greenspan, H., et al. (eds.) International Conference on Medical Image Computing and Computer-Assisted Intervention, vol. 14228, pp. 101–111. Springer, Cham (2023). https://doi.org/10.1007/978-3-031-43996-4_10
Yoon, J., Lee, J., Heo, S., et al.: hSDB-instrument: Instrument localization database for laparoscopic and robotic surgeries. In: de Bruijne, M., et al. (eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2021, pp. 393–402. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87202-1_38
Yu, W., Si, C., Zhou, P., et al.: MetaFormer baselines for vision. IEEE Trans. Pattern Anal. Mach. Intell. 46(2), 896–912 (2024). https://doi.org/10.1109/tpami.2023.3329173
Zhang, Y., Bano, S., Page, A.S., Deprest, J., Stoyanov, D., Vasconcelos, F.: Retrieval of surgical phase transitions using reinforcement learning. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022, pp. 497–506. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-16449-1_47
Zia, A., Bhattacharyya, K., Liu, X., et al.: Surgical tool classification and localization: results and methods from the MICCAI 2022 SurgToolLoc challenge (2023)
Acknowledgements
We thank SURF (www.surf.nl) for the support in using the National Supercomputer Snellius.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2025 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Jaspers, T.J.M. et al. (2025). Exploring the Effect of Dataset Diversity in Self-supervised Learning for Surgical Computer Vision. In: Bhattarai, B., et al. Data Engineering in Medical Imaging. DEMI 2024. Lecture Notes in Computer Science, vol 15265. Springer, Cham. https://doi.org/10.1007/978-3-031-73748-0_5
Download citation
DOI: https://doi.org/10.1007/978-3-031-73748-0_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-73747-3
Online ISBN: 978-3-031-73748-0
eBook Packages: Computer ScienceComputer Science (R0)