Abstract
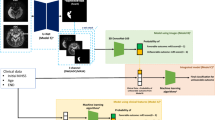
Accurate functional outcome prediction shortly after the onset of stroke would enable more effective personalized care of stroke patients. A deep learning approach was developed that utilizes follow-up images at 24 h to predict the functional outcome 90 days after stroke onset. The method involves the use of a conventional U-net segmentation model that was trained to delineate the stroke lesion on CT images, with an additional branch integrated into the U-net’s bottom layer to extract features for a separately trained network that classifies cases into favorable (modified Rankin Score (mRS) = 0–2) or unfavorable outcomes (mRS = 3–6). The method was trained and validated using 3-fold cross-validation on a set of 240 images (training: 170; validation: 90). The lesion segmentation yielded an average Dice score of 0.458 and an average absolute volume error of 10.71 ml, while the binary mRS prediction obtained an overall accuracy of 88.6% on the training set and 58.0% on the validation set. Although there is much room for improvement, these results demonstrate the potential of deep learning approaches toward a promising decision-support tool for stroke outcome prediction.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Stroke. Mayo Clinic. https://www.mayoclinic.org/diseases-conditions/stroke/symptoms-causes/syc-20350113, last checked on 12 May 2023, https://www.mayoclinic.org/diseases-conditions/stroke/symptoms-causes/syc-20350113
Bacchi, S., Zerner, T., Oakden-Rayner, L., Kleinig, T., Patel, S., Jannes, J.: Deep learning in the prediction of ischaemic stroke thrombolysis functional outcomes: a pilot study. Acad. Radiol. 27, e19–e23 (2020). https://doi.org/10.1016/j.acra.2019.03.015
Banks, J.L., Marotta, C.A.: Outcomes validity and reliability of the modified rankin scale: implications for stroke clinical trials - a literature review and synthesis. Stroke 38, 1091–1096 (2007). https://doi.org/10.1161/01.STR.0000258355.23810.c6
Bertels, J., Robben, D., Lemmens, R., Vandermeulen, D.: DeepVoxNet2: yet another CNN framework, November 2022. http://arxiv.org/abs/2211.09569
Choi, Y., Kwon, Y., Lee, H., Kim, B.J., Paik, M.C., Won, J.H.: Ensemble of deep convolutional neural networks for prognosis of ischemic stroke, pp. 231–243 (2016)
Ding, L., Liu, Z., Mane, R., Wang, S., Jing, J., et al., H.F.: Predicting functional outcome in patients with acute brainstem infarction using deep neuroimaging features. Eur. J. Neurol. 29, 744–752 (2022). https://doi.org/10.1111/ene.15181
Feigin, V.L., Brainin, M., Norrving, B., Martins, S., Sacco, R.L., et al., W.H.: World stroke organization (WSO): global stroke fact sheet 2022, November 2022. https://doi.org/10.1177/17474930211065917
Goyal, M., Ospel, J.M., Kappelhof, M., Ganesh, A.: Challenges of outcome prediction for acute stroke treatment decisions. Stroke 1921–1928 (2021). https://doi.org/10.1161/STROKEAHA.120.033785
Heylen, E., Vandermeulen, D., Maes, F., Dupont, P., Scheldeman, L., Lambert, J., Bertels, J.: Modelling the evolution of brain tissue status in ischemic stroke using CNNs. Master thesis, KU Leuven (2021)
Karolinska University Hospital. https://www.karolinska.se/en/karolinska-university-hospital/
Lin, C.H., Hsu, K.C., Johnson, K.R., Fann, Y.C., Tsai, C.H., et al., Y.S.: Evaluation of machine learning methods to stroke outcome prediction using a nationwide disease registry. Comput. Methods Program. Biomed. 190 (2020). https://doi.org/10.1016/j.cmpb.2020.105381
Nielsen, A., Hansen, M.B., Tietze, A., Mouridsen, K.: Prediction of tissue outcome and assessment of treatment effect in acute ischemic stroke using deep learning. Stroke 49, 1394–1401 (2018). https://doi.org/10.1161/STROKEAHA.117.019740
Nishi, H., et al.: Deep learning-derived high-level neuroimaging features predict clinical outcomes for large vessel occlusion. Stroke 1484–1492 (2020). https://doi.org/10.1161/STROKEAHA.119.028101
Omarov, B., et al.: Modified unet model for brain stroke lesion segmentation on computed tomography images. Comput. Mater. Continua 71, 4701–4717 (2022). https://doi.org/10.32604/cmc.2022.020998
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation, pp. 234–241 (2015)
Zhou, Y., Huang, W., Dong, P., Xia, Y., Wang, S.: D-unet: a dimension-fusion u shape network for chronic stroke lesion segmentation, August 2019. https://doi.org/10.1109/TCBB.2019.2939522, http://arxiv.org/abs/1908.05104
Acknowledgements
We want to express our sincere gratitude to Fredrik Ståhl and Åke Holmberg for providing the KAROLINSKA dataset, which has been instrumental in facilitating our research endeavors.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Ulens, M. et al. (2024). From Brain Tissue Infarction at 24 Hours to Patient Functional Outcome at 90 Days Using Deep Learning. In: Baid, U., et al. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes SWITCH 2023 2023. Lecture Notes in Computer Science, vol 14668. Springer, Cham. https://doi.org/10.1007/978-3-031-76160-7_11
Download citation
DOI: https://doi.org/10.1007/978-3-031-76160-7_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-76159-1
Online ISBN: 978-3-031-76160-7
eBook Packages: Computer ScienceComputer Science (R0)