Abstract
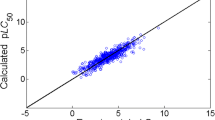
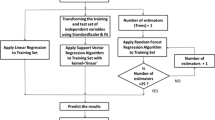
As there has been a rise in the usage of in silico approaches, for assessing the risks of harmful chemicals upon animals, more researchers focus on the utilization of Quantitative Structure Activity Relationship models. A number of machine learning algorithms link molecular descriptors that can infer chemical structural properties associated with their corresponding biological activity. Efficient and comprehensive computational methods which can process huge set of heterogeneous chemical datasets are in demand. In this context, this study establishes the usage of various machine learning algorithms in predicting the acute aquatic toxicity of diverse chemicals on Fathead Minnow (Pimephales promelas). Sample drive approach is employed on the train set for binning the data so that they can be located in a domain space having more similar chemicals, instead of using the dataset that covers a wide range of chemicals at the entirety. Here, bin wise best learning model and subset of features that are minimally required for the classification are found for further ease. Several regression methods are employed to find the estimation of toxicity LC50 value by adopting several statistical measures and hence bin wise strategies are determined. Through experimentation, it is evident that the proposed model surpasses the other existing models by providing an R2 of 0.8473 with RMSE 0.3035 which is comparable. Hence, the proposed model is competent for estimating the toxicity in new and unseen chemical.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Cassotti, M., Ballabio, D., Todeschini, R., Consonni, V.: A similarity-based QSAR model for predicting acute toxicity towards the fathead minnow (Pimephalespromelas). SAR QSAR Environ. Res. 26(3), 217–243 (2015)
In, Y., Lee, S.K., Kim, P.J., No, K.T.: Prediction of acute toxicity to fathead minnow by local model based QSAR and global QSAR approaches. Bull. Korean Chem. Soc. 33(2), 613–619 (2012)
Devillers, J.: A new strategy for using supervised artificial neural networks in QSAR. SAR QSAR Environ. Res. 16(5), 433–442 (2005)
Sheffield, T.Y., Judson, R.S.: Ensemble QSAR modeling to predict multispecies fish toxicity lethal concentrations and points of departure. Environ. Sci. Technol. 53, 12793−12802 (2019)
Gajewicz-Skretna, A., Furuhama, A., Yamamoto, H., Suzuki, N.: Generating accurate in silico predictions of acute aquatic toxicity for a range of organic chemicals: towards similarity-based machine learning methods. Chemosphere 280, 130681 (2021)
Karim, A., et al.: Quantitative toxicity prediction via meta ensembling of multitask deep learning models. ACS Omega 6, 12306−12317 (2021)
Singh, K.P., Gupta, S., Kumar, A., Mohan, D.: Multispecies QSAR modeling for predicting the aquatic toxicity of diverse organic chemicals for regulatory toxicology. Chem. Res. Toxicol. 27, 741−753 (2014)
Nendzat, M., Russomi, C.L.: QSAR modelling of the ERL-D fathead minnow acute toxicity database. Xenobiotica 27(2), 147–170 (1991)
Wang, Y., Chen, X.: A joint optimization QSAR model of fathead minnow acute toxicity based on a radial basis function neural network and its consensus modeling. RSC Adv. 10, 21292 (2020)
Liu, H., Setiono, R.: Chi2: feature selection and discretization of numeric attributes. In: 7th IEEE International Conference Proceedings on Tools with Artificial Intelligence, pp. 388–391 (1995)
Lozano, S., Lescot, E., Halm, M.-P., Lepailleur, A., Bureau, R., Rault, S.: Prediction of acute toxicity in fish by using QSAR methods and chemical modes of action. J. Enzyme Inhibit. Med. Chem. 25(2), 195–203 (2010)
Dearden, J.C., Cronin, M.T.D., Kaiser, K.L.E.: How not to develop a quantitative structure–activity or structure–property relationship (QSAR/QSPR). SAR QSAR Environ. Res. 20(3–4), 241–266 (2009)
Rakhimbekova, A., et al.: Cross-validation strategies in QSPR modelling of chemical reactions. SAR QSAR Environ. Res. 32(3), 207–219 (2021)
Lovrić, M., Malev, O., Klobučar, G., Kern, R., Liu, J.J., Lučić, B.: Predictive capability of QSAR models based on the CompTox zebrafish embryo assays: an imbalanced classification problem. Molecules 26, 1617 (2021)
Judson, R.: ToxValDB: Compiling Publicly Available In Vivo Toxicity Data. Presented at EPA’s Computational Toxicology Communities of Practice Monthly Meeting, RTP, NC, (2018)
Cassotti, M., Ballabio, D., Consonni, V., Mauri, A., Tetko, I.V., Todeschini, R.: Prediction of acute aquatic toxicity towards Daphnia magna by using the GA-kNN method. ATLA-Alternatives to Laboratory Animals 42, 31–41 (2014)
Enoch, S.J., Cronin, M.T.D., Schultz, T.W., Madden, J.C.: An evaluation of global QSAR models for the prediction of the toxicity of phenols to Tetrahymena pyriformis. Chemosphere 71, 1225–1232 (2008)
Toma, C., Cappelli, C.I., Manganaro, A., Lombardo, A., Arning, J., Benfenati, E.: New models to predict the acute and chronic toxicities of representative species of the main trophic levels of aquatic environments. Molecules 26, 6983 (2021)
Wu, X., Zhang, Q., Hu, J.: QSAR study of the acute toxicity to fathead minnow based on a large dataset. SAR QSAR Environ. Res. 27(2), 147–164 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2025 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Kavitha, R., Guru, D.S. (2025). Quantitative Structure-Activity Relationship Modeling for the Prediction of Fish Toxicity Lethal Concentration on Fathead Minnow. In: Antonacopoulos, A., Chaudhuri, S., Chellappa, R., Liu, CL., Bhattacharya, S., Pal, U. (eds) Pattern Recognition. ICPR 2024. Lecture Notes in Computer Science, vol 15310. Springer, Cham. https://doi.org/10.1007/978-3-031-78192-6_3
Download citation
DOI: https://doi.org/10.1007/978-3-031-78192-6_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-78191-9
Online ISBN: 978-3-031-78192-6
eBook Packages: Computer ScienceComputer Science (R0)