Abstract
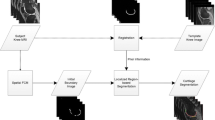
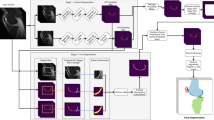
Atlas based segmentation techniques have been proven to be effective in many automatic segmentation applications. However, the reliance on image correspondence means that the segmentation results can be affected by any registration errors which occur, particularly if there is a high degree of anatomical variability. This paper presents a novel multi-resolution patch-based segmentation framework which is able to work on images without requiring registration. Additionally, an image similarity metric using 3D histograms of oriented gradients is proposed to enable atlas selection in this context. We applied the proposed approach to segment MR images of the knee from the MICCAI SKI10 Grand Challenge, where 100 training atlases are provided and evaluation is conducted on 50 unseen test images. The proposed method achieved good scores overall and is comparable to the top entries in the challenge for cartilage segmentation, demonstrating good performance when comparing against state-of-the-art approaches customised to Knee MRI.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Akgül, C.B., Rubin, D.L., Napel, S., Beaulieu, C.F., Greenspan, H., Acar, B.: Content-Based Image Retrieval in Radiology: Current Status and Future Directions. Journal of Digital Imaging 24(2), 208–222 (2011)
Aljabar, P., Heckemann, R.A., Hammers, A., Hajnal, J.V., Rueckert, D.: Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. NeuroImage 46(3), 726–738 (2009)
Anderson, C.H., Bergen, J.R., Burt, P.J., Ogden, J.M.: Pyramid Methods in Image Processing (1984)
Coupé, P., Manjón, J.V., Fonov, V., Pruessner, J., Robles, M., Collins, D.L.: Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. NeuroImage 54(2), 940–954 (2011)
Donoghue, C.R., Rao, A., Pizarro, L., Bull, A.M.J., Rueckert, D.: Fast and accurate global geodesic registrations using knee MRI from the Osteoarthritis Initiative. In: CVPRW, pp. 50–57 (June 2012)
Eskildsen, S.F., Coupé, P., Fonov, V., Manjón, J.V., Leung, K.K., Guizard, N., Wassef, S.N., Østergaard, L.R., Collins, D.L.: BEaST: brain extraction based on nonlocal segmentation technique. NeuroImage 59(3), 2362–2373 (2012)
Fripp, J., Crozier, S., Warfield, S.K., Ourselin, S.: Automatic Segmentation and Quantitative Analysis of the Articular Cartilages from Magnetic Resonance Images of the Knee. IEEE Transactions on Medical Imaging 29(1), 55–64 (2010)
Heckemann, R.A., Hajnal, J.V., Aljabar, P., Rueckert, D., Hammers, A.: Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage 33(1), 115–126 (2006)
Heimann, T., Morrison, B.: Segmentation of knee images: A grand challenge. In: MICCAI Workshop on Medical Image Analysis for the Clinic, pp. 207–214 (2010)
Klaeser, A., Marszalek, M., Schmid, C.: A Spatio-Temporal Descriptor Based on 3D-Gradients. In: Procedings of BMVC, pp. 99.1–99.10 (2008)
Maurer, C.R., Rensheng, Q., Raghavan, V.: A linear time algorithm for computing exact euclidean distance transforms of binary images in arbitrary dimensions. IEEE Transactions on Pattern Analysis and Machine Intelligence 25(2), 265–270 (2003)
Nyúl, L.G., Udupa, J.K.: On standardizing the MR image intensity scale. Magnetic Resonance in Medicine 42(6), 1072–1081 (1999)
Rohlfing, T., Brandt, R., Menzel, R., Maurer, C.R.: Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. NeuroImage 21(4), 1428–1442 (2004)
Rousseau, F., Habas, P., Studholme, C.: A Supervised Patch-Based Approach for Human Brain Labeling. IEEE Transactions on Medical Imaging 30(10), 1852–1862 (2011)
Thomson, J.: On the Structure of the Atom: an Investigation of the Stability and Periods of Oscillation of a number of Corpuscles arranged at equal intervals around the Circumference of a Circle; with Application of the Results to the Theory of Atomic Structure. Philosophical Magazine 7(39), 237–265 (1904)
Torralba, A., Fergus, R., Freeman, W.T.: 80 Million Tiny Images: a Large Data Set for Nonparametric Object and Scene Recognition. IEEE Transactions on Pattern Analysis and Machine Intelligence 30(11), 1958–1970 (2008)
Tustison, N.J., Avants, B.B., Cook, P.A., Zheng, Y., Egan, A., Yushkevich, P.A., Gee, J.C.: N4ITK: improved N3 bias correction. IEEE Transactions on Medical Imaging 29(6), 1310–1320 (2010)
Wang, Z., Wolz, R., Tong, T., Rueckert, D.: Spatially Aware Patch-based Segmentation (SAPS): An Alternative Patch-Based Segmentation Framework. In: Menze, B.H., Langs, G., Lu, L., Montillo, A., Tu, Z., Criminisi, A. (eds.) MCV 2012. LNCS, vol. 7766, pp. 93–103. Springer, Heidelberg (2013)
Wolz, R., Chu, C., Misawa, K., Mori, K., Rueckert, D.: Multi-organ Abdominal CT Segmentation Using Hierarchically Weighted Subject-Specific Atlases. In: Ayache, N., Delingette, H., Golland, P., Mori, K. (eds.) MICCAI 2012, Part I. LNCS, vol. 7510, pp. 10–17. Springer, Heidelberg (2012)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer International Publishing Switzerland
About this paper
Cite this paper
Wang, Z., Donoghue, C., Rueckert, D. (2013). Patch-Based Segmentation without Registration: Application to Knee MRI. In: Wu, G., Zhang, D., Shen, D., Yan, P., Suzuki, K., Wang, F. (eds) Machine Learning in Medical Imaging. MLMI 2013. Lecture Notes in Computer Science, vol 8184. Springer, Cham. https://doi.org/10.1007/978-3-319-02267-3_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-02267-3_13
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-02266-6
Online ISBN: 978-3-319-02267-3
eBook Packages: Computer ScienceComputer Science (R0)