Abstract
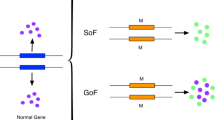
Genomic variations have been intensively studied since the development of high-throughput sequencing technologies. There are numerous tools and databases predicting and annotating the functional impact of genetic variants, such as determining whether a variant is neutral or deleterious to the functions of the corresponding protein. However, there is a need for methods that not only identify neutral or deleterious mutations but also provide fine grained prediction on the outcome resulting from mutations, such as gain, loss, or switch of function. This paper proposes the deployment of multiple hidden Markov models to computationally classify mutations by functional impact type.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Pauline, C., Henikoff, S.: Predicting Deleterious amino acid substitutions. Genome Res. 111, 863–874 (2001)
Ramensky, V., Bork, P., Sunyaev, S.: Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30(17), 3894–3900 (2002)
Cooper, G., Stone, E., Asimenos, G.: Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15(7), 901–913 (2005)
Asthana, S., Roytberg, M., Stamatoyannopoulos, J.: Analysis of sequence conservation at nucleotide resolution. PLOS Comput. Biol. 3, e254 (2007)
Reva, B., Antipin, Y., Sander, C.: Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118 (2011)
Lee, W., et al.: Bi-directional SIFT predicts a subset of activating mutations. PLoS ONE 4, e8311 (2009)
Ng, S., et al.: PARADIGM-SHIFT predicts the function of mutations in multiple cancers using pathway impact analysis. Bioinformatics 28, i640–i646 (2012)
Liu, M., Watson, L.T., Zhang, L.: Quantitative prediction of the effect of genetic variation using hidden Markov models. BMC Bioinformatics 15, 5 (2014)
Edgar, R.C.: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 32(5), 1792–1797 (2004)
Petitjean, A., Mathe, E., Kato, S.: Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 28, 622–629 (2007)
Kato, S., Han, S., Liu, W.: Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. U.S.A. 100(14), 8424–8429 (2003)
Breiman, L.: Random Forests. Machine Learning 45(1), 5–32 (2001)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this paper
Cite this paper
Liu, M., Watson, L.T., Zhang, L. (2014). Classification of Mutations by Functional Impact Type: Gain of Function, Loss of Function, and Switch of Function. In: Basu, M., Pan, Y., Wang, J. (eds) Bioinformatics Research and Applications. ISBRA 2014. Lecture Notes in Computer Science(), vol 8492. Springer, Cham. https://doi.org/10.1007/978-3-319-08171-7_21
Download citation
DOI: https://doi.org/10.1007/978-3-319-08171-7_21
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-08170-0
Online ISBN: 978-3-319-08171-7
eBook Packages: Computer ScienceComputer Science (R0)