Abstract
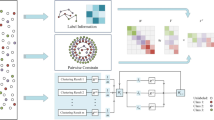
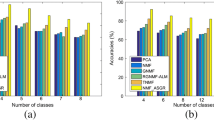
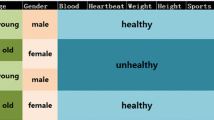
Determination of the appropriate number of clusters is a big challenge for the bi-clustering method of the non-negative matrix factorization (NMF). The conventional determination method may be to test a number of candidates and select the optimal one with the best clustering performance. However, such strategy of repetition test is obviously time-consuming. In this paper, we propose a novel efficient algorithm called the automatic NMF clustering method with competitive sparseness constraints (autoNMF) which can perform the reasonable clustering without pre-assigning the exact number of clusters. It is demonstrated by the experiments that the autoNMF has been significantly improved on both clustering performance and computational efficiency.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Kim, H., Park, H.: Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics 23, 1495–1502 (2007)
Brunet, J.-P., Tamayo, P., Golub, T.R., Mesirov, J.P.: Metagenes and molecular pattern discovery using matrix factorization. Proceedings of the National Academy of Sciences 101, 4164–4169 (2004)
Lawson, C.L., Hanson, R.J.: Solving least squares problems, vol. 161. SIAM (1974)
Kim, H., Park, H.: Nonnegative matrix factorization based on alternating nonnegativity constrained least squares and active set method. SIAM Journal on Matrix Analysis and Applications 30, 713–730 (2008)
Peharz, R., Pernkopf, F.: Sparse nonnegative matrix factorization with ℓ0-constraints. Neurocomputing 80, 38–46 (2012)
Lee, D.D., Seung, H.S.: Algorithms for Non-Negative Matrix Factorization. In: Advances in Neural Information Processing Systems, pp. 556–562 (2000)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this paper
Cite this paper
Liu, C., Ma, J. (2014). Automatic Non-negative Matrix Factorization Clustering with Competitive Sparseness Constraints. In: Huang, DS., Jo, KH., Wang, L. (eds) Intelligent Computing Methodologies. ICIC 2014. Lecture Notes in Computer Science(), vol 8589. Springer, Cham. https://doi.org/10.1007/978-3-319-09339-0_12
Download citation
DOI: https://doi.org/10.1007/978-3-319-09339-0_12
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-09338-3
Online ISBN: 978-3-319-09339-0
eBook Packages: Computer ScienceComputer Science (R0)