Abstract
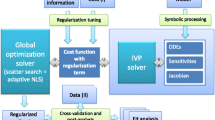
Kinetic models are being increasingly used as a systematic framework to understand function in biological systems. Calibration of these nonlinear dynamic models remains challenging due to the nonconvexity and ill-conditioning of the associated inverse problems. Nonconvexity can be dealt with suitable global optimization. Here, we focus on simultaneously dealing with ill-conditioning by making use of proper regularization methods. Regularized calibrations ensure the best trade-offs between bias and variance, thus reducing over-fitting. We present a critical comparison of several methods, and guidelines for properly tuning them. The performance of this procedure and its advantages are illustrated with a well known benchmark problem considering several scenarios of data availability and measurement noise.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Almquist, J., Cvijovic, M., Hatzimanikatis, V., Nielsen, J., Jirstrand, M.: Kinetic models in industrial biotechnology - Improving cell factory performance. Metabolic Engineering 1–22 (April 2014)
Ashyraliyev, M., Fomekong-Nanfack, Y., Kaandorp, J.A., Blom, J.G.: Systems biology: parameter estimation for biochemical models. FEBS Journal 276(4), 886–902 (2009)
Banga, J.R., Balsa-Canto, E.: Parameter estimation and optimal experimental design. Essays in Biochemistry 45, 195–210 (2008)
Bansal, L., Chu, Y., Laird, C., Hahn, J.: Regularization of Inverse Problems to Determine Transcription Factor Profiles from Fluorescent Reporter Systems. AIChE Journal 58(12), 3751–3762 (2012)
Bauer, F.: Some considerations concerning regularization and parameter choice algorithms. Inverse Problems 23(2), 837–858 (2007)
Bauer, F., Lukas, M.A.: Comparingparameter choice methods for regularization of ill-posed problems. Mathematics and Computers in Simulation 81(9), 1795–1841 (2011)
Dennis, J.E., Gay, D.M., Welsch, R.E.: An Adaptive Nonlinear Least-Squares Algorithm. ACM Transaction on Mathematical Software 7(3), 348–368 (1981)
Engl, H.W., Flamm, C., Kügler, P., Lu, J., Müller, S., Schuster, P., Philipp, K.: Inverse problems in systems biology. Inverse Problems 25(12), 123014 (2009)
Engl, H.W., Hanke, M., Neubauer, A.: Regularization of Inverse Problems. Kluwer Academic Publishers (1996)
Hansen, P.C., O’Leary, D.P.: The use of the L-Curve in the regularization of discrete ill-posed problems. SIAM Journal on Scientific Computing 14(6), 1487–1503 (1993)
Hindmarsh, A.C., Brown, P.N., Grant, K.E., Lee, S.L., Serban, R., Shumaker, D.E., Woodward, C.S.: {SUNDIALS: Suite of Nonlinear and Differential / Algebraic Equation Solvers}. ACM Transaction on Mathematical Software 31(3), 363–396 (2005)
Kaltenbacher, B., Neubauer, A., Scherzer, O.: Iterative Regularization Methods for Nonlinear Ill-Posed Problems. Radon Series on Computational and Applied Mathematics. Walter de Gruyter, Berlin, New York (2008)
Kitano, H. (ed.): Foundations of Systems Biology. The MIT Press (2001)
Kravaris, C., Hahn, J., Chu, Y.: Advances and selected recent developments in state and parameter estimation. Computers & Chemical Engineering 51, 111–123 (2013), http://linkinghub.elsevier.com/retrieve/pii/S0098135412001779
Lepskii, O.: On a Problem of Adaptive Estimation in Gaussian White Noise. Theory of Probability & Its Applications 35(3), 454–466 (1991)
Link, H., Christodoulou, D., Sauer, U.: Advancing metabolic models with kinetic information. Current Opinion in Biotechnology 29, 8–14 (2014)
Ljung, L., Chen, T.: What can regularization offer for estimation of dynamical systems? In: 11th IFAC International Workshop on Adaptation and Learning in Control and Signal Processing. IFAC, vol. 5 (2013)
Miró, A., Pozo, C., Guillén-Gosálbez, G., Egea, J.A., Jiménez, L.: Deterministic global optimization algorithm based on outer approximation for the parameter estimation of nonlinear dynamic biological systems. BMC Bioinformatics 13(1), 90 (2012)
Moles, C.G., Mendes, P., Banga, J.R.: Parameter Estimation in Biochemical Pathways: A Comparison of Global Optimization Methods. Genome Research 13, 2467–2474 (2003)
Morozov, V.A.: Methods for Solving Incorrectly Posed Problems. Springer (1984)
Palm, R.: Numerical Comparison of Regularization Algorithms for Solving Ill-Posed Problems. Ph.D. thesis, University of Tartu, Estonia
Papamichail, I., Adjiman, C.S.: Global optimization of dynamic systems. Comput. Chem. Eng. 28, 403–415 (2004)
Regiska, T.: A Regularization Parameter in Discrete Ill-Posed Problems. SIAM Journal on Scientific Computing 17(3), 740–749 (1996)
Reichel, L., Rodriguez, G.: Old and new parameter choice rules for discrete ill-posed problems. Numerical Algorithms 63(1), 65–87 (2012)
Rodriguez-Fernandez, M., Egea, J.A., Banga, J.R.: Novel metaheuristic for parameter estimation in nonlinear dynamic biological systems. BMC Bioinformatics 7, 483 (2006)
Rodriguez-Fernandez, M., Mendes, P., Banga, J.R.: A hybrid approach for efficient and robust parameter estimation in biochemical pathways. Bio Systems 83(2-3), 248–265 (2006)
Schmidt, M., Fung, G., Rosaless, R.: Optimization Methods for l1-Regularization. Tech. rep. (2009), http://www.cs.ubc.ca/cgi-bin/tr/2009/TR-2009-19.pdf
Stanford, N.J., Lubitz, T., Smallbone, K., Klipp, E., Mendes, P., Liebermeister, W.: Systematic construction of kinetic models from genome-scale metabolic networks. PloS one 8(11), e79195 (2012)
Tibshirani, R.: Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 58(1), 267–288 (1996)
Walter, E., Prorizato, L.: Identification of Parametric Models from experimental data. Springer (1997)
Wang, H., Wang, X.C.: Parameter estimation for metabolic networks with two stage Bregman regularization homotopy inversion algorithm. Journal of Theoretical Biology 343, 199–207 (2014)
Zou, H., Hastie, T.: Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 67(2), 301–320 (2005), http://doi.wiley.com/10.1111/j.1467-9868.2005.00503.x
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this paper
Cite this paper
Gábor, A., Banga, J.R. (2014). Improved Parameter Estimation in Kinetic Models: Selection and Tuning of Regularization Methods. In: Mendes, P., Dada, J.O., Smallbone, K. (eds) Computational Methods in Systems Biology. CMSB 2014. Lecture Notes in Computer Science(), vol 8859. Springer, Cham. https://doi.org/10.1007/978-3-319-12982-2_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-12982-2_4
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-12981-5
Online ISBN: 978-3-319-12982-2
eBook Packages: Computer ScienceComputer Science (R0)