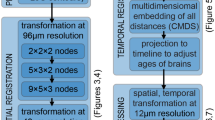
Abstract
Atlases of the human brain have numerous applications in neurological imaging such as the analysis of brain growth. Publicly available atlases of the developing brain have previously been constructed using the arithmetic mean of free-form deformations which were obtained by asymmetric pairwise registration of brain images. Most of these atlases represent cross-sections of the growth process only. In this work, we use the Log-Euclidean mean of inverse consistent transformations which belong to the one-parameter subgroup of diffeomorphisms, as it more naturally represents average morphology. During the registration, similarity is evaluated symmetrically for the images to be aligned. As both images are equally affected by the deformation and interpolation, asymmetric bias is reduced. We further propose to represent longitudinal change by exploiting the numerous transformations computed during the atlas construction in order to derive a deformation model of mean growth. Based on brain images of 118 neonates, we constructed an atlas which describes the dynamics of early development through mean images at weekly intervals and a continuous spatio-temporal deformation. The evolution of brain volumes calculated on preterm neonates is in agreement with recently published findings based on measures of cortical folding of fetuses at the equivalent age range.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Aljabar, P., Bhatia, K.K., Hajnal, J.V., Boardman, J.P., Srinivasan, L., Rutherford, M.A., Dyet, L.E., Edwards, A.D., Rueckert, D.: Analysis of growth in the developing brain using non-rigid registration. In: ISBI, pp. 201–204 (2006)
Arsigny, V., Commowick, O., Pennec, X., Ayache, N.: A log-euclidean framework for statistics on diffeomorphisms. In: Larsen, R., Nielsen, M., Sporring, J. (eds.) MICCAI 2006. LNCS, vol. 4190, pp. 924–931. Springer, Heidelberg (2006)
Avants, B.B., Epstein, C.L., Grossman, M., Gee, J.C.: Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12(1), 26–41 (2008)
Bossa, M., Hernandez, M., Olmos, S.: Contributions to 3D diffeomorphic atlas estimation: application to brain images. In: Ayache, N., Ourselin, S., Maeder, A. (eds.) MICCAI 2007, Part I. LNCS, vol. 4791, pp. 667–674. Springer, Heidelberg (2007)
Cheng, S.H.U.N., Higham, N.J., Kenney, C.S., Laub, A.J.: Approximating the logarithm of a matrix to specified accuracy. SIAM J. Matrix Anal. Appl. 22(4), 1112–1125 (1999)
Davis, B.C., Fletcher, P.T., Bullitt, E., Joshi, S.: Population shape regression from random design data. In: ICCV, pp. 1–7 (2007)
Habas, P.A., Kim, K., Corbett-Detig, J.M., Rousseau, F., Glenn, O.A., Barkovich, A.J., Studholme, C.: A spatiotemporal atlas of MR intensity, tissue probability and shape of the fetal brain with application to segmentation. NeuroImage 53(2), 460–470 (2010)
Joshi, S., Davis, B., Jomier, M., Gerig, G.: Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage 23(suppl. 1), S151–S160 (2004)
Kuklisova-Murgasova, M., Aljabar, P., Srinivasan, L., Counsell, S.J., Doria, V., Serag, A., Gousias, I.S., Boardman, J.P., Rutherford, M.A., Edwards, A.D., Hajnal, J.V., Rueckert, D.: A dynamic 4D probabilistic atlas of the developing brain. NeuroImage 54(4), 2750–2763 (2011)
Ledig, C., Wright, R., Serag, A., Aljabar, P., Rueckert, D.: Neonatal brain segmentation using second order neighborhood information. In: Workshop on Perinatal and Paediatric Imaging: PaPI, MICCAI, pp. 33–40 (2012)
Lorenzi, M., Ayache, N., Frisoni, G.B., Pennec, X.: LCC-Demons: a robust and accurate symmetric diffeomorphic registration algorithm. NeuroImage 81, 470–483 (2013)
Modat, M., Cardoso, M.J., Daga, P., Cash, D., Fox, N.C., Ourselin, S.: Inverse-consistent symmetric free form deformation. In: Dawant, B.M., Christensen, G.E., Fitzpatrick, J.M., Rueckert, D. (eds.) WBIR 2012. LNCS, vol. 7359, pp. 79–88. Springer, Heidelberg (2012)
Modat, M., Daga, P., Cardoso, M.J., Ourselin, S., Ridgway, G.R., Ashburner, J.: Parametric non-rigid registration using a stationary velocity field. In: MMBIA, pp. 145–150 (2012)
Modat, M., Ridgway, G.R., Taylor, Z.A., Hawkes, D.J., Fox, N.C., Ourselin, S.: A parallel-friendly normalised mutual information gradient for free-form registration. In: SPIE (2009)
Niethammer, M., Huang, Y., Vialard, F.-X.: Geodesic regression for image time-series. In: Fichtinger, G., Martel, A., Peters, T. (eds.) MICCAI 2011, Part II. LNCS, vol. 6892, pp. 655–662. Springer, Heidelberg (2011)
Pluim, J.P.W., Maintz, J.B.A., Viergever, M.A.: Mutual-information-based registration of medical images: a survey. IEEE TMI 22(8), 986–1004 (2003)
Rueckert, D., Sonoda, L.I., Hayes, C., Hill, D.L., Leach, M.O., Hawkes, D.J.: Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18(8), 712–721 (1999)
Serag, A., Aljabar, P., Ball, G., Counsell, S.J., Boardman, J.P., Rutherford, M.A., Edwards, A.D., Hajnal, J.V., Rueckert, D.: Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. NeuroImage 59(3), 2255–2265 (2012)
Singh, N., Hinkle, J., Joshi, S., Fletcher, P.T.: A vector momenta formulation of diffeomorphisms for improved geodesic regression and atlas construction. In: ISBI, pp. 1219–1222 (2013)
Vercauteren, T., Pennec, X., Perchant, A., Ayache, N.: Symmetric log-domain diffeomorphic registration: a demons-based approach. In: Metaxas, D., Axel, L., Fichtinger, G., Székely, G. (eds.) MICCAI 2008, Part I. LNCS, vol. 5241, pp. 754–761. Springer, Heidelberg (2008)
Wright, R., Kyriakopoulou, V., Ledig, C., Rutherford, M.A., Hajnal, J.V., Rueckert, D., Aljabar, P.: Automatic quantification of normal cortical folding patterns from fetal brain MRI. NeuroImage 91, 21–32 (2014)
Yushkevich, P.A., Avants, B.B., Das, S.R., Pluta, J., Altinay, M., Craige, C.: Bias in estimation of hippocampal atrophy using deformation-based morphometry arises from asymmetric global normalization: an illustration in ADNI 3T MRI data. NeuroImage 50(2), 434–445 (2010)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Schuh, A. et al. (2015). Construction of a 4D Brain Atlas and Growth Model Using Diffeomorphic Registration. In: Durrleman, S., Fletcher, T., Gerig, G., Niethammer, M., Pennec, X. (eds) Spatio-temporal Image Analysis for Longitudinal and Time-Series Image Data. STIA 2014. Lecture Notes in Computer Science(), vol 8682. Springer, Cham. https://doi.org/10.1007/978-3-319-14905-9_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-14905-9_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-14904-2
Online ISBN: 978-3-319-14905-9
eBook Packages: Computer ScienceComputer Science (R0)