Abstract
3D medical image segmentation is needed for diagnosis and treatment. As manual segmentation is very costly, automatic segmentation algorithms are needed. For finding best algorithms, several algorithms need to be evaluated on a set of organ instances. This is currently difficult due to dataset size and complexity.
In this paper, we present a novel method for comparison and evaluation of several algorithms that automatically segment 3D medical images. It combines algorithmic data analysis with interactive data visualization. A clustering algorithm identifies regions of common quality across the segmented data set for each algorithm. The comparison identifies best algorithms per region. Interactive views show the algorithm quality.
We applied our approach to a real-world cochlea dataset, which was segmented with several algorithms. Our approach allowed segmentation experts to compare algorithms on regional level and to identify best algorithms per region.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In medicine, detection of organs and organic structures in 3D images is an important task during diagnosis and treatment. Manual segmentation is very expensive as it requires expertise and is very time consuming. Therefore, automatic segmentation algorithms are needed. The development of these algorithms is difficult due to image resolution, image noisiness and organ variation.
The development of segmentation algorithms requires a detailed evaluation of segmentation results and their comparison across various algorithms or algorithm variations. The evaluation of segmentation quality often takes a set of ground truth images (i.e., expert segmentations) and compares them to automatic segmentation results. Then the segmentation quality is compared across algorithms (see Fig. 1). It is important to identify where algorithms systematically fail and to identify best performing algorithms. This needs to be done on a local (per point or region) level, as some algorithms may perform better on one part of the organ (e.g., top) and at the same time fail in other region (e.g. bottom). In these cases, an expert needs to know in which part of the organ, which algorithm is better suitable and how well it performs.
Current algorithm evaluation and comparison methods support the algorithm assessment only in a limited way. Often one remains at a global evaluation, which means that for each mesh a score is created that evaluates the algorithmic segmentation. Since this only gives a rough overview of the quality, it does not allow to see whether the algorithms have problems in specific regions. In contrary, local evaluation methods require a detailed inspection of each individual result, without the possibility to compare the results algorithmically. A visual comparison is in this case limited to several instances due to screen size. Moreover, it is time consuming and subjective. So there is a need for new methods allowing for detailed comparison of segmentation quality for several algorithms.
In this paper, we present a novel method for comparison and evaluation of several 3D medical image segmentation algorithms. Our approach combines algorithmic data analysis with interactive data visualization. A specialized clustering algorithm identifies regions of common quality across data set for each algorithm. It thereby allows for comparison of segmentation algorithms on local level. The comparison identifies best algorithms per region. Interactive views show the algorithm quality on various levels of detail. We concentrate on algorithms based on statistical shape models. These algorithms are widely used owing to their robustness and good quality.
We applied our approach to a real-world cochlea dataset which was segmented with several algorithms. Our approach allowed segmentation experts to compare algorithms on regional level and to identify best algorithms per region.
The paper is structured as follows. Section 2 presents related work in this area. Section 3 details on our approach. Section 5 concludes and outlines future work.
2 Related Work
We review related publications. We first review standard 3D segmentation evaluation methods used by medical imaging experts. We then focus on visual analytics methods for the evaluation of SSM-based segmentations. As our focus is not on segmentation, we note that review of SSM based segmentation methods can be found in [9].
Currently, medical image segmentation experts mainly employ two evaluation methods: algorithmic and visual. Algorithmic evaluation relies on a set of global metrics [8]. The metrics include Average Surface Distance, Maximum Surface Distance or Dice Coefficient. The global measures provide only a coarse information on segmentation quality. They do not discriminate between two results which are badly segmented in different regions (e.g., top and bottom of an organ). Visual inspection concerns individual instances shown in 2D and 3D views such as ITK Snap (www.itksnap.org) or overlay the two meshes using transparency, color-coded local distances between vertices, e.g., [2, 5, 6, 13]. These views are very detailed, but require manual inspection of all instances individually. They are not suitable for several algorithms with multiple instances.
Visual Analysis for evaluation of segmentation results and comparative visualization is the topic of several recent works reviewed in [7, 11]. Some works analyze the effect of segmentation parameters on the result (e.g., [14]). They do not compare several instances for several algorithm results, which is in the focus of our work. Other works focus on shape variability analysis [3, 10] without evaluating segmentation quality.
Cardness et al. [4] presented an interactive visualization system for analyzing segmentation quality. Local segmentation quality view is combined with the calculation of global segmentation quality metrics. This approach, however, does not enable quality comparison. Von Landesberger et al. [11] presented several methods for supporting creation and evaluation of medical image segmentation algorithms. They show the distribution of global quality values across the dataset and select instances with high or low quality values for detailed inspection. This visualization compares only global quality values, it does not allow for comparing local quality across the dataset. A recent publication [15] analyzes the progress of segmentation quality during the segmentation process. It analyzes segmentation quality for one organ in each segmentation iteration. This approach allows for analyzing and comparing local quality improvements, but is constrained to one sample. Quality comparison across samples or across algorithms is not possible.
3 Approach
We have developed an approach that consists of two types of visual evaluations supported by quality-based clustering (see Fig. 2):
-
Global Quality View: The data are globally evaluated in a scatterplot matrix. It offers pairwise comparison of algorithm’s quality pro instance. It is based on global quality criteria. It also allows to select instances for further regional analysis.
-
Quality-based Clustering: Quality-based clustering identifies regions with systematic quality characteristics across the test dataset. Clusters are then further processed in order to determine algorithms with best output quality pro region.
-
Regional Quality View: Regional view shows the clustering and algorithm quality results.
Overview of our approach. First, global quality comparison in a scatterplot matrix is performed. It offers pairwise comparison of algorithm quality pro instance. Then clustering is performed to identify regions with systematic quality characteristics and to identify best algorithms pro region. The results are explored in regional view.
Global Quality View: This view allows the user to compare algorithms for each instance in a set (see Fig. 2). It provides first insights into an overall algorithm quality and enables their comparison across algorithms. The comparison employs common global segmentation quality measures, such as Hausdorff Distance and Average Surface distance [8].
Global Quality view shows a scatterplot matrix of algorithm comparisons (see Fig. 3). The scatterplots show pairwise comparisons of algorithms, where the points are segmentation instances (see Fig. 4). Upper right matrix shows comparison for average surface distance, lower left triangle shows results for Hausdorff distance. Diagonal shows result quality pro algorithm. In each scatterplot, each X-axis represents quality of one algorithm, and the Y axis shows the quality of another algorithm. The scatterplot allows the user to assess the quality of each segmentation (bottom left corner - good, upper right corner - bad). It also allows to analyze which algorithm is better for a particular instance.
The user can use this view also for selecting instances for further regional analysis. For example, the user can exclude outliers from further analysis.
Explanation of global comparison view. The scatterplots show pairwise comparisons of algorithms, where the points are segmentation instances. Upper right matrix shows comparison for average surface distance, lower left triangle shows results for Hausdorff distance. Diagonal shows result quality pro algorithm. In each scatterplot, each X-axis represents quality of one algorithm, and the Y axis shows the quality of another algorithm.
Quality-based Clustering and identification of best algorithms: Landmarks of an organ are clustered according to their quality values so that regions with similar quality appear. We use a connectivity-extended hierarchical agglomerative clustering. After the regions are detected, we automatically identify which algorithms are best for each region. Suitable algorithms are those with low Average surface distance in a region. As several algorithms can lead to very good and similar results, we use a user-defined quality tolerance level for best algorithms. For example, all algorithms with values less than the best algorithm value + tolerance are deemed suitable for a particular region.
Regional Quality View: The regional quality comparison view allows the user to gain an insight into which algorithms are suitable for which region of an organ. It shows the results of a previous algorithmic analysis.
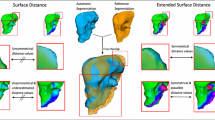
The regional comparison view shows the identified regions using a black border (see Fig. 5 left). Within each region, the best algorithm is indicated by a color. If several algorithms are suitable for a region, then they are displayed using weaving of colors within this region, as inspired by [12]. In this way, the segmentation experts can see which algorithms perform best on the organ and in which region.
For a detailed quality assessment, the user can select a region and explore quality values in an additional view (see Fig. 5 right). This view shows the quality values of all landmarks within a region for all suitable algorithms. In this way, the user can both assess the segmentation quality and compare the algorithm qualities.
4 Application
Segmentation experts used our approach for comparing the quality of several segmentation algorithms for cochlea. The input dataset consists of 20 cochlea instances gained from a CT Scanner with an average intraslice voxel size of 0.18 and an average interslice voxel size of 0.38 mm. All instances have a manually created ground truth segmentation. Automatic segmentations have been generated using three variations of the SSM-based algorithm by Becker et al. [1].
Global view (see Fig. 3) shows that average distance is lower than 0.3 mm, thus the samples are very well segmented. All algorithms have broadly similar result quality. As the samples (i.e., points) are close to the diagonal in most scatterplots, there seem to be no large differences among algorithms. Looking at the individual samples shows that samples 5 and 9 (S5 and S9) have the best segmentation results. Interestingly, Sample 4 (S4) has extraordinary bad quality according to Hausdorff distance for all algorithms, but it has “normal” quality according to average distance. This indicates that this sample has good quality on average, but has some badly segmented regions (see Fig. 6).
Regional view shows the regions on cochlea with similar quality characteristics. The clustering algorithms identified 21 regions on cochlea. The largest region is very well segmented by all three algorithms. It has a mean quality of 0.075 mm (see Fig. 5 bottom). The algorithm could also identify a very well segmented region in the middle of cochlea. However, solely the Algorithms 2 and 3 have been determined as the best in this region (see Fig. 5 top).
5 Conclusions and Future Work
We presented a new system for visual comparison of several segmentation algorithms on a dataset containing multiple 3D images. Our approach allows the user to analyze and visualize the quality of segmentation algorithms according to local quality.
We applied our approach to a cochlea dataset segmented with three versions of a SSM-based algorithm. Segmentation experts were able to assess the quality of algorithms on a dataset.
In the future, we would like to extend the scalability of our approach.
References
Becker, M., Kirschner, M., Sakas, G.: Segmentation of risk structures for otologic surgery using the probabilistic active shape model (PASM), 9036, 90360O–90360O-7 (2014)
Busking, S., Botha, C.P., Ferrarini, L., Milles, J., Post, F.H.: Image-based rendering of intersecting surfaces for dynamic comparative visualization. Vis. Comput. 27(5), 347–363 (2011)
Busking, S., Botha, C.P., Post, F.H.: Dynamic multi-view exploration of shape spaces. Comput. Graph. Forum 29(3), 973–982 (2010)
Cárdenes, R., Bach, M., Chi, Y., Marras, I., de Luis, R., Anderson, M., Cashman, P., Bultelle, M.: Multimodal evaluation for medical image segmentation. In: Kropatsch, W.G., Kampel, M., Hanbury, A. (eds.) CAIP 2007. LNCS, vol. 4673, pp. 229–236. Springer, Heidelberg (2007)
Dick, C., Burgkart, R., Westermann, R.: Distance visualization for interactive 3d implant planning. IEEE Trans. Vis. Comput. Graph. 17(12), 2173–2182 (2011)
Gerig, Guido, Jomier, Matthieu, Chakos, Miranda: Valmet: A New validation tool for assessing and improving 3D object segmentation. In: Niessen, Wiro J., Viergever, Max A. (eds.) MICCAI 2001. LNCS, vol. 2208, p. 516. Springer, Heidelberg (2001)
Gleicher, M., Albers, D., Walker, R., Jusufi, I., Hansen, C.D., Roberts, J.C.: Visual comparison for information visualization. Inf. Vis. 10(4), 289–309 (2011)
Heimann, T., van Ginneken, B., Styner, M., et al.: Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans. Med. Imaging 28, 1251–1265 (2009)
Heimann, T., Meinzer, H.P.: Statistical shape models for 3D medical image segmentation: A review. Med. Image Anal. 13(4), 543–563 (2009)
Klemm, P., Lawonn, K., Rak, M., Preim, B., Toennies, K.D., Hegenscheid, K., Völzke, H., Oeltze, S.: Visualization and analysis of lumbar spine canal variability in cohort study data. In: Proceedings of the International Workshop on Vision, Modeling and Visualization, pp. 121–128 (2013)
von Landesberger, T., Bremm, S., Kirschner, M., Wesarg, S., Kuijper, A.: Visual analytics for model-based medical image segmentation: Opportunities and challenges. Expert Syst. Appl. 40(12), 4934–4943 (2013)
Luboschik, M., Radloff, A., Schumann, H.: A new weaving technique for handling overlapping regions. In: Proceedings of the International Conference on Advanced Visual Interfaces, AVI 2010, pp. 25–32. ACM, New York (2010). http://doi.acm.org/10.1145/1842993.1842999
Schmidt, J., Preiner, R., Auzinger, T., Wimmer, M., Gröller, M.E., Bruckner, S.: Ymca - your mesh comparison application. In: IEEE VIS 2014. IEEE Computer Society, Nov 2014
Torsney-Weir, T., Saad, A., Moller, T., Hege, H.C., Weber, B., Verbavatz, J., Bergner, S.: Tuner: principled parameter finding for image segmentation algorithms using visual response surface exploration. IEEE Trans. Vis. Comput. Graph. 17(12), 1892–1901 (2011)
von Landesberger, T., Andrienko, G., Andrienko, N., Bremm, S., Kirschner, M., Wesarg, S., Kuijper, A.: Opening up the black box of medical image segmentation with statistical shape models. Vis. Comput. 29(9), 893–905 (2013). doi:10.1007/s00371-013-0852-y
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Geurts, A., Sakas, G., Kuijper, A., Becker, M., Landesberger, T.v. (2015). Visual Comparison of 3D Medical Image Segmentation Algorithms Based on Statistical Shape Models. In: Duffy, V. (eds) Digital Human Modeling. Applications in Health, Safety, Ergonomics and Risk Management: Ergonomics and Health. DHM 2015. Lecture Notes in Computer Science(), vol 9185. Springer, Cham. https://doi.org/10.1007/978-3-319-21070-4_34
Download citation
DOI: https://doi.org/10.1007/978-3-319-21070-4_34
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-21069-8
Online ISBN: 978-3-319-21070-4
eBook Packages: Computer ScienceComputer Science (R0)