Abstract
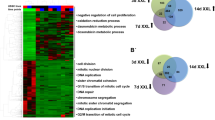
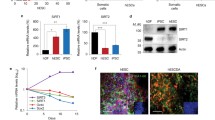
Elucidating the role that microRNAs (miRNAs) and signaling transduction play in the directed differentiation of human embryonic stem cells (hESCs) into glucose-responsive, insulin-producing endocrine cells is critical to our understanding of systems biology and the development of cell-based therapeutics. To accomplish this, a biochemical understanding the underpinnings of hESC differentiation bias – the propensity of hESCs to differentiate into cells of a specific lineage – must be described in molecular detail. An inherent aspect of hESC culture is stress, and we hypothesize that stress is largely responsible for differentiation bias. Our results indicate that manipulating stress increases apoptosis and disrupts differentiation. Cells subjected to stress fail to become endocrine precursor cells and retain many characteristics of pluripotent cells. Many stresses induce massive apoptosis and result in a loss of up to 80 % of the cells. A consequence of the reduction in cell density is elevated stress signaling, dramatic changes in cell proliferation, maintenance of pluripotency markers, and a complete absence of transcription factors associated with pancreatic endocrine cell production. Coincident with changes in stress, we observed dramatic changes in correlated miRNAexpression, suggesting that cell stress may modulate miRNA transcription and ultimately hESC differentiation.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bagga, S., et al.: Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563 (2005)
Bartel, D.P.: MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004)
Hinton, A., Hunter, S., Reyes, G., Fogel, G.B., King, C.C.: From pluripotency to islets: miRNAs as critical regulators of human cellular differentiation. Adv. Genet. 79, 1–34 (2012)
Bagga, S., Pasquinelli, A.E.: Identification and analysis of microRNAs. Genet. Eng. (N Y) 27, 1–20 (2006)
Ritchie, W., Rasko, J.E., Flamant, S.: MicroRNA target prediction and validation. Adv. Exp. Med. Biol. 774, 39–53 (2013)
Wang, Y., Medvid, R., Melton, C., Jaenisch, R., Blelloch, R.: DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380–385 (2007)
Suh, M.R., et al.: Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270, 488–498 (2004)
Suh, N., et al.: MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr. Biol. 20, 271–277 (2010)
Lynn, F.C., et al.: MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes 56, 2938–2945 (2007)
Hinton, A., et al.: A distinct microRNA signature for definitive endoderm derived from human embryonic stem cells. Stem Cells Dev. 19, 797–807 (2010)
Hinton, A., et al.: sRNA-seq analysis of human embryonic stem cells and definitive endoderm reveal differentially expressed microRNAs and novel isomiRs with distinct targets. Stem Cells 32, 2360–2372 (2014)
Remondini, D., Neretti, N., Franceschi, C., Tieri, P., Sedivy, J.M., Milanesi, L., Castellani, G.C.: Networks from gene expression time series: characterization of correlation patterns. Int. J. Bifurcat. Chaos 17(7), 2477–2483 (2007)
Acknowledgements
Funding for these studies was provided by the Larry L. Hillblom Foundation (CCK) and the California Institute for Regenerative Medicine (CIRM to CCK). The authors would also like to thank Sevan Ficici for his assistance.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Fogel, G.B., Tallon, T., Wong, A.S., Lopez, A.D., King, C.C. (2015). miRNA Regulation of Human Embryonic Stem Cell Differentiation. In: Lones, M., Tyrrell, A., Smith, S., Fogel, G. (eds) Information Processing in Cells and Tissues. IPCAT 2015. Lecture Notes in Computer Science(), vol 9303. Springer, Cham. https://doi.org/10.1007/978-3-319-23108-2_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-23108-2_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-23107-5
Online ISBN: 978-3-319-23108-2
eBook Packages: Computer ScienceComputer Science (R0)