Abstract
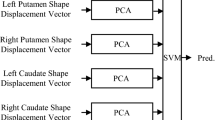
This work introduces a robust framework for predicting Deep Brain Stimulation (DBS) target structures which are not identifiable on standard clinical MRI. While recent high-field MR imaging allows clear visualization of DBS target structures, such high-fields are not clinically available, and therefore DBS targeting needs to be performed on the standard clinical low contrast data. We first learn via regression models the shape relationships between DBS targets and their potential predictors from high-field (7 Tesla) MR training sets. A bagging procedure is utilized in the regression model, reducing the variability of learned dependencies. Then, given manually or automatically detected predictors on the clinical patient data, the target structure is predicted using the learned high quality information. Moreover, we derive a robust way to properly weight different training subsets, yielding higher accuracy when using an ensemble of predictions. The subthalamic nucleus (STN), the most common DBS target for Parkinson’s disease, is used to exemplify within our framework. Experimental validation from Parkinson’s patients shows that the proposed approach enables reliable prediction of the STN from the clinical 1.5T MR data.
Chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
References
Abosch, A., Yacoub, E., Ugurbil, K., Harel, N.: An assessment of current brain targets for deep brain stimulation surgery with susceptibility weighted imaging at 7 tesla. Neurosurgery 67(6), 1745–1756 (2010)
Daniluk, S., Davies, K.G., Ellias, S.A., Novak, P., Nazzaro, J.M.: Asssessment of the variability in the anatomical position and size of the subthalamic nucleus among patients with advanced Parkinsons disease using magnetic resonance imaging. Acta Neurochirurgica 152(2), 201–210 (2010)
Xiao, Y., Jannin, P., D’Albis, T., Guizard, N., Haegelen, C., Lalys, F., Vérin, M., Collins, D.L.: Investigation of Morphometric Variability of Subthalamic Nucleus, Red Nucleus, and Substantia Nigra in Advanced Parkinsons Disease Patients Using Automatic Segmentation and PCA-Based Analysis. Human Brain Mapping 35(9), 4330–4344 (2014)
Lenglet, C., Abosch, A., Yacoub, E., De Martino, F., Sapiro, G., Harel, N.: Comprehensive in vivo mapping of the human basal ganglia and thalamic connectome in individuals using 7T MRI. PLoS One 7, e29153 (2012)
Kim, J., Lenglet, C., Duchin, Y., Sapiro, G., Harel, N.: Semiautomatic segmentation of brain subcortical structures from high-field MRI. IEEE J. Biomed. Health Inform. 18(5), 1678–1695 (2014)
Cho, Z., Min, H., Oh, S., Han, J., Park, C., Chi, J., Kim, Y., Paek, S.H., Lozano, A.M., Lee, K.H.: Direct visualization of deep brain stimulation targets in Parkinson disease with the use of 7-tesla magnetic resonance imaging. J. Neurosurg. 113(3), 639–647 (2010)
Rao, A., Aljabar, P., Rueckert, D.: Hierarchical statistical shape analysis and prediction of sub-cortical brain structures. Med. Image Anal. 12(1), 55–68 (2008)
Baka, N., Metz, C., Schaap, M., Lelieveldt, B., Niessen, W., De Bruijne, M.: Comparison of shape regression methods under landmark position uncertainty. In: Fichtinger, G., Martel, A., Peters, T. (eds.) MICCAI 2011, Part II. LNCS, vol. 6892, pp. 434–441. Springer, Heidelberg (2011)
Blanc, R., Seiler, C., Szekely, G., Nolte, L., Reyes, M.: Statistical model based shape prediction from a combination of direct observations and various surrogates: Application to orthopaedic research. Med. Image Anal. 16(6), 1156–1166 (2012)
Criminisi, A., Robertson, D., Konukoglu, E., Shotton, J., Pathak, S., White, S., Siddiqui, K.: Regression forests for efficient anatomy detection and localization in computed tomography scans. Med. Image Anal. 17(8), 1293–1303 (2013)
Breiman, L.: Bagging predictors. Machine Learning 24(2), 123–140 (1996)
Duchin, Y., Abosch, A., Yacoub, E., Sapiro, G., Harel, N.: Feasibility of using ultra-high field (7T) MRI for clinical surgical targeting. PLoS One 7, e37328 (2012)
Avants, B.B., Tustison, N.J., Song, G., Cook, P.A., Klein, A., Gee, J.C.: A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54(3), 2033–2044 (2011)
Heimann, T., Wolf, I., Williams, T., Meinzer, H.: 3d active shape models using gradient descent optimization of description length. In: Christensen, G.E., Sonka, M. (eds.) IPMI 2005. LNCS, vol. 3565, pp. 566–577. Springer, Heidelberg (2005)
Cootes, T.F., Taylor, C.J., Cooper, D.H., Graham, J.: Active shape models – Their training and application. Comput. Vis. Image Understand 61(1), 38–59 (1995)
Rathi, Y., Dambreville, S., Tannenbaum, A.: Statistical shape analysis using kernel PCA. In: SPIE-IS&T Electronic Imaging (2010)
Abdi, H.: Partial least squares regression and projection on latent structure regression (PLS Regression). Wiley Interdiscip. Rev. Comp. Stat. 2(1), 97–106 (2010)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Kim, J., Duchin, Y., Kim, H., Vitek, J., Harel, N., Sapiro, G. (2015). Robust Prediction of Clinical Deep Brain Stimulation Target Structures via the Estimation of Influential High-Field MR Atlases. In: Navab, N., Hornegger, J., Wells, W., Frangi, A. (eds) Medical Image Computing and Computer-Assisted Intervention -- MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science(), vol 9350. Springer, Cham. https://doi.org/10.1007/978-3-319-24571-3_70
Download citation
DOI: https://doi.org/10.1007/978-3-319-24571-3_70
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-24570-6
Online ISBN: 978-3-319-24571-3
eBook Packages: Computer ScienceComputer Science (R0)