Abstract
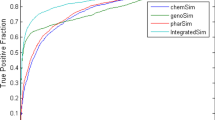
Prediction of drug-drug interactions (DDIs) is an essential step in both drug development and clinical application. As the number of approved drugs increases, the number of potential DDIs rapidly rises. Several drugs have been withdrawn from the market due to DDI-related adverse drug reactions recently. Therefore, it is necessary to develop an accurate prediction tool that can identify potential DDIs during clinical trials. We propose a new methodology for DDIs prediction by integrating the drug-drug pair similarity, including drug phenotypic, therapeutic, structural, and genomic similarity. A large-scale study was conducted to predict 6946 known DDIs of 721 approved drugs. The area under the receiver operating characteristic curve of the integrated models is 0.953 as evaluated using five-fold cross-validation. Additionally, the integrated model is able to detect the biological effect produced by the DDI. Through the integration of drug phenotypic, therapeutic, structural, and genomic similarities, we demonstrated that the proposed method is simple, efficient, allows the uncovering DDIs in the drug development process and postmarketing surveillance.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Beijnen, J.H., Schellens, J.H.: Drug interactions in oncology. Lancet Oncol. 5, 489–496 (2004)
Nemeroff, C.B., Preskorn, S.H., Devane, C.L.: Antidepressant drug-drug interactions: clinical relevance and risk management. CNS Spectr. 12, 1–13 (2007)
Classen, D.C., Pestotnik, S.L., Evans, R.S., Lloyd, J.F., Burke, J.P.: Adverse drug events in hospitalized patientsExcess length of stay, extra costs, and attributable mortality. JAMA 277, 301–306 (1997)
Gottlieb, S.: Antihistamine drug withdrawn by manufacturer. BMJ. Br. Med. J. 319, 7 (1999)
Henney, J.E.: Withdrawal of troglitazone and cisapride. JAMA, J. Am. Med. Assoc. 283, 2228 (2000)
SoRelle, R.: Withdrawal of Posicor from market. Circulation 98, 831–832 (1998)
Moore, T.J., Cohen, M.R., Furberg, C.D.: Serious adverse drug events reported to the food and drug administration, 1998–2005. Arch. Intern. Med. 167, 1752–1759 (2007)
Bjornsson, T.D., Callaghan, J.T., Einolf, H.J., Fischer, V., Gan, L., et al.: The conduct of in vitro and in vivo drug-drug interaction studies: a pharmaceutical research and manufacturers of America (PhRMA) perspective. Drug Metab. Dispos. 31, 815–832 (2003)
Cheng, F., Li, W., Liu, G., Tang, Y.: In silico ADMET prediction: recent advances, current challenges and future trends. Curr. Top. Med. Chem. 13, 1273–1289 (2013)
Percha, B., Altman, R.B.: Informatics confronts drug–drug interactions. Trends Pharmacol. Sci. 34, 178–184 (2013)
Gottlieb, A., Stein, G.Y., Oron, Y., Ruppin, E., Sharan, R.: INDI: a computational framework for inferring drug interactions and their associated recommendations. Mol. Syst. Biol. 8, 592 (2012)
Huang, J., Niu, C., Green, C.D., Yang, L., Mei, H., et al.: Systematic prediction of pharmacodynamic drug-drug interactions through protein-protein-interaction network. PLoS Comput. Biol. 9, e1002998 (2013)
Cami, A., Manzi, S., Arnold, A., Reis, B.Y.: Pharmacointeraction network models predict unknown drug-drug interactions. PLoS ONE 8, e61468 (2013)
Cheng, F., Zhao, Z.: Machine learning-based prediction of drug–drug interactions by integrating drug phenotypic, therapeutic, chemical, and genomic properties. J. Am. Med. Inform. Assoc. 21, e278–e286 (2014)
Vilar, S., Harpaz, R, Uriarte, E., Santana, L., Rabadan., R., et al.: Drug–drug interaction through molecular structure similarity analysis. J. Am. Med. Inform. Assoc. (2012). doi:10.1136/amiajnl-2012-000935
Wishart, D.S., Knox, C., Guo, A.C., et al.: DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucl. Acids Res. 36(suppl 1), D901–D906 (2008)
Knox, C., Law, V., Jewison, T., Liu, P., Ly, S., et al.: DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucl. Acids Res. 39, D1035–D1041 (2011)
OLBoyle, N.M., Banck, M., James, C.A., Morley, C., Vandermeersch, T., et al.: Open babel: an open chemical toolbox. J. Cheminf. 3, 33 (2011)
Cheng, F., Li, W., Wu, Z., Wang, X., Zhang, C., et al.: Prediction of polypharmacological profiles of drugs by the integration of chemical, side effect, and therapeutic space. J. Chem. Inf. Model. 53, 753–762 (2013)
Xu, K.-J., Song, J., Zhao, X.-M.: The drug cocktail network. BMC Syst. Biol. 6, S5 (2012)
Zhu, F., Shi, Z., Qin, C., Tao, L., Liu, X., et al.: Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucl. Acids Res. 40, D1128–D1136 (2012)
Willett, P.: Similarity-based virtual screening using 2D fingerprints. Drug Discov. Today 11, 1046–1053 (2006)
Vilar, S., Karpiak, J., Costanzi, S.: Ligand and structure-based models for the prediction of ligand-receptor affinities and virtual screenings: development and application to the β2-adrenergic receptor. J. Comput. Chem. 31, 707–720 (2010)
Engel, S., Skoumbourdis, A.P., Childress, J., Neumann, S., Deschamps, J.R., et al.: A virtual screen for diverse ligands: discovery of selective G protein-coupled receptor antagonists. J. Am. Chem. Soc. 130, 5115–5123 (2008)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Wang, B., Yu, X., Wei, R., Yuan, C., Li, X., Zheng, CH. (2016). System Prediction of Drug-Drug Interactions Through the Integration of Drug Phenotypic, Therapeutic, Structural, and Genomic Similarities. In: Huang, DS., Bevilacqua, V., Premaratne, P. (eds) Intelligent Computing Theories and Application. ICIC 2016. Lecture Notes in Computer Science(), vol 9771. Springer, Cham. https://doi.org/10.1007/978-3-319-42291-6_37
Download citation
DOI: https://doi.org/10.1007/978-3-319-42291-6_37
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-42290-9
Online ISBN: 978-3-319-42291-6
eBook Packages: Computer ScienceComputer Science (R0)