Abstract
In Foot-and-Mouth Disease Virus (FMDV), understanding how viruses offer protection against related emerging strains is vital for creating effective vaccines. With testing large numbers of vaccines being infeasible, the development of an in silico predictor of cross-protection between virus strains has been a vital area of recent research. The current paper reviews a recent contribution to this area, the SABRE method, a sparse hierarchical Bayesian model which uses spike and slab priors to identify key antigenic sites within FMDV serotypes. WAIC is then combined with the SABRE method and its ability to approximate Bayesian Cross Validation performance in terms of correctly selecting random effect components analysed. WAIC and the SABRE method have then been applied to two FMDV datasets and the results analysed.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Andrieu, C., Doucet, A.: Joint Bayesian model selection and estimation of noisy sinusoids via reversible jump MCMC. IEEE Trans. Sig. Process. 47(10), 2667–2676 (1999)
Bates, D., Maechler, M., Bolker, B.: lme4: linear mixed-effects models using S4 classes (2013)
Davies, V., Reeve, R., Harvey, W., Maree, F., Husmeier, D.: Sparse Bayesian variable selection for the identification of antigenic variability in the Foot-and-Mouth Disease Virus. J. Mach. Learn. Res. Workshop Conf. Proc. (AISTATS) 33, 149–158 (2014)
Gelman, A., Carlin, J.B., Stern, H.S., Dunson, D.B., Ventari, A., Rubin, D.B.: Bayesian Data Analysis, 3rd edn. Chapman & Hall, London (2013)
Gelman, A., Rubin, D.: Inference from iterative simulation using multiple sequences. Stat. Sci. 7, 457–511 (1992)
George, E.I., McCulloch, R.E.: Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 88(423), 881–889 (1993)
Grazioli, S., Moretti, M., Barbieri, I., Crosatti, M., Brocchi, E.: Use of monoclonal antibodies to identify and map new antigenic determinants involved in neutralisation on FMD viruses type SAT 1 and SAT 2. In: Report of the Session of the Research Group of the Standing Technical Committee of the European Commission for the Control of Foot-and-Mouth Disease, pp. 287–297, Appendix 43 (2006)
Harvey, W.T., Gregory, V., Benton, D.J., Hall, J.P., Daniels, R.S., Bedford, T., Haydon, D.T., Hay, A.J., McCauley, J.W., Reeve, R.: Identifying the genetic basis of antigenic change in influenza A (H1N1). arXiv preprint arXiv:1404.4197 (2015)
Hastie, T., Tibshirani, R., Friedman, J.: The Elements of Statistical Learning. Springer, New York (2009)
Hastings, W.: Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57(1), 97–109 (1970)
Jow, H., Boys, R.J., Wilkinson, D.J.: Bayesian identification of protein differential expression in multi-group isobaric labelled mass spectrometry data. Stat. Appl. Genet. Mol. Biol. 13(5), 531–551 (2014)
Maree, F.F., Borley, D.W., Reeve, R., Upadhyaya, S., Lukhwareni, A., Mlingo, T., Esterhuysen, J.J., Harvey, W.T., Fry, E.E., Parida, S., Paton, D.J., Mahapatra, M.: Tracking the antigenic evolution of Foot-and-Mouth Disease Virus (2015, in submission)
Metropolis, N., Rosenbluth, A., Rosenbluth, M., Teller, A., Teller, E.: Equations of state calculations by fast computing machines. J. Chem. Phys. 21(6), 1087–1092 (1953)
Mitchell, T., Beauchamp, J.: Bayesian variable selection in linear regression. J. Am. Stat. Assoc. 83(404), 1023–1032 (1988)
Mohamed, S., Heller, K., Ghahramani, Z.: Bayesian and \(l_1\) approaches for sparse unsupervised learning. In: Proceedings of the 29th International Conference on Machine Learning (ICML 2012), pp. 751–758 (2012)
Pinheiro, J.C., Bates, D.: Mixed-Effects Models in S and S-PLUS. Springer, New York (2000)
Reeve, R., Blignaut, B., Esterhuysen, J.J., Opperman, P., Matthews, L., Fry, E.E., de Beer, T.A.P., Theron, J., Rieder, E., Vosloo, W., O’Neill, H.G., Haydon, D.T., Maree, F.F.: Sequence-based prediction for vaccine strain selection and identification of antigenic variability in Foot-and-Mouth Disease Virus. PLoS Comput. Biol. 6(12), e1001027 (2010)
Schelldorfer, J., Bühlmann, P., van de Geer, S.: Estimation for high-dimensional linear mixed-effects models using \({\ell }1\)-penalization. Scand. J. Stat. 38(2), 197–214 (2011)
Tibshirani, R.: Regression shrinkage and selection via the lasso. J. Roy. Stat. Soc. B 58, 267–288 (1996)
Watanabe, S.: Asymptotic equivalence of Bayes cross validation and widely applicable information criterion in singular learning theory. J. Mach. Learn. Res. 11, 3571–3594 (2010)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
10 Appendix
10 Appendix
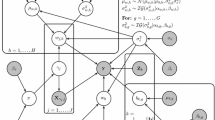
For the Gibbs sampling we sample the intercept and regression coefficients together and define \(\mathbf w _{\varvec{\gamma }}^* = (w_0,\mathbf w _{\varvec{\gamma }}^\top )^\top \), \(\mathbf X _{\varvec{\gamma }}^* = (\mathbf 1 ,\mathbf X _{\varvec{\gamma }})\), \(\mathbf m _{\varvec{\gamma }} = (\mu _{w_0},\mu _{w,1},\ldots ,\mu _{w,1},\) \(\mu _{w,2},\ldots ,\mu _{w,H})^\top \) and \(\varvec{\varSigma }_\mathbf{w _{\varvec{\gamma }}^*} = diag(\varvec{\sigma }_\mathbf{w ^*}^2)\) with \(\varvec{\sigma }_\mathbf{w ^*}^2 = (\sigma ^2_{w_0},\sigma ^2_{w,1},\ldots ,\sigma ^2_{w,1},\sigma ^2_{w,2},\) \(\ldots ,\sigma ^2_{w,H})^\top \). Each \(\mu _{w,h}\) and \(\sigma _{w,h}^2\) is repeated with length \(||\mathbf w _{\varvec{\gamma },h}||\) dependent on \(\varvec{\gamma }\). The Gibbs sampling distributions are then given as follows, with \(\varvec{\theta }'\) used to denote all the parameters not on the left of the conditioning bar:
where we sample \(\sigma _{b,g}^2\), \(\mu _{w,h}\) and \(\sigma _{w,h}^2\) for each g and h respectively. We also define \(\mathbf V _\mathbf{w _{\varvec{\gamma }}^*} = (\mathbf X _{\varvec{\gamma }}^{*\top } \mathbf X _{\varvec{\gamma }}^* + \varvec{\varSigma }_\mathbf{w _{\varvec{\gamma }}^*}^{-1})^{-1}\), \(\mathbf V _\mathbf{b } = (\tfrac{1}{\sigma _\varepsilon ^2 }{} \mathbf Z ^\top \mathbf Z + \varvec{\varSigma }_\mathbf{b }^{-1})^{-1}\), \(V_{\mu _\gamma ,h} = ((||\mathbf w _{\varvec{\gamma },h}|| / \sigma _{w,h}^{2})^{-1}\) \(+ (\sigma _{0,h}^2)^{-1})^{-1}\) and \(R_{\sigma _\varepsilon ^2} = (\mathbf y - \mathbf X _{\varvec{\gamma }}^* \mathbf w _{\varvec{\gamma }}^* -\mathbf Z {} \mathbf b )^\top (\mathbf y - \mathbf X _{\varvec{\gamma }}^* \mathbf w _{\varvec{\gamma }}^* -\mathbf Z {} \mathbf b ) + (\mathbf w _{\varvec{\gamma }}^* - \mathbf m _{\varvec{\gamma }})^\top \varvec{\varSigma }_\mathbf{w _{\varvec{\gamma }}^*}^{-1}\) \((\mathbf w _{\varvec{\gamma }}^*\) \(- \mathbf m _{\varvec{\gamma }}) + \sum _{h=1}^H (\mu _{w,h} - \mu _{0,h})^2 / \sigma _{0,h}^2\) for notational simplicity.
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Davies, V., Reeve, R., Harvey, W.T., Husmeier, D. (2016). Selecting Random Effect Components in a Sparse Hierarchical Bayesian Model for Identifying Antigenic Variability. In: Angelini, C., Rancoita, P., Rovetta, S. (eds) Computational Intelligence Methods for Bioinformatics and Biostatistics. CIBB 2015. Lecture Notes in Computer Science(), vol 9874. Springer, Cham. https://doi.org/10.1007/978-3-319-44332-4_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-44332-4_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-44331-7
Online ISBN: 978-3-319-44332-4
eBook Packages: Computer ScienceComputer Science (R0)