Abstract
Understanding brain volumetry is essential to understand neuro-development and disease. Historically, age-related changes have been studied in detail for specific age ranges (e.g., early childhood, teen, young adults, elderly, etc.) or more sparsely sampled for wider considerations of lifetime aging. Recent advancements in data sharing and robust processing have made available considerable quantities of brain images from normal, healthy volunteers. However, existing analysis approaches have had difficulty addressing (1) complex volumetric developments on the large cohort across the life time (e.g., beyond cubic age trends), (2) accounting for confound effects, and (3) maintaining an analysis framework consistent with the general linear model (GLM) approach pervasive in neuroscience. To address these challenges, we propose to use covariate-adjusted restricted cubic spline (C-RCS) regression within a multi-site cross-sectional framework. This model allows for flexible consideration of nonlinear age-associated patterns while accounting for traditional covariates and interaction effects. As a demonstration of this approach on lifetime brain aging, we derive normative volumetric trajectories and 95 % confidence intervals from 5111 healthy patients from 64 sites while accounting for confounding sex, intracranial volume and field strength effects. The volumetric results are shown to be consistent with traditional studies that have explored more limited age ranges using single-site analyses. This work represents the first integration of C-RCS with neuroimaging and the derivation of structural covariance networks (SCNs) from a large study of multi-site, cross-sectional data.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Hierarchical Cluster Analysis
- Total Intracranial Volume
- Label Fusion
- General Linear Model Framework
- Whole Brain Volume
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Brain volumetry across the lifespan is essential in neurological research and clinical investigation. Magnetic resonance imaging (MRI) allows for quantification of such changes, and consequent investigation of specific age ranges or more sparsely sampled lifetime data [1]. Contemporaneous advancements in data sharing have made considerable quantities of brain images available from normal, healthy populations. However, the regression models prevalent in volumetric mapping (e.g., linear, polynomial, non-parametric model, etc.) have had difficulty in modeling complex, cross-sectional large cohorts while accounting for confound effects.
This paper proposes a novel multi-site cross-sectional framework using Covariate-adjusted Restricted Cubic Spline (C-RCS) regression to map brain volumetry on a large cohort (5111 MR 3D images) across the lifespan (4 ~ 98 years). The C-RCS extends the Restricted Cubic Spline [2, 3] by regressing out the confound effects in a general linear model (GLM) fashion. Multi-atlas segmentation is used to obtain whole brain volume (WBV) and 132 regional volumes. The regional volumes are further grouped to 15 networks of interest (NOIs). Then, structural covariance networks (SCNs), i.e. regions or networks that mature or decline together during developmental periods, are established based on NOIs using hierarchical clustering analysis (HCA). To validate the large-scale framework, confidence intervals (CI) are provided for both C-RCS regression and clustering from 10,000 bootstrap samples.
2 Methods
2.1 Extracting Volumetric Information
The complete cohort aggregates 9 datasets with a total 5111 MR T1w 3D images from normal healthy subjects (Table 1). 45 atlases are non-rigidly registered [4] to a target image and non-local spatial staple (NLSS) label fusion [5] is used to fuse the labels from each atlas to the target image using the BrainCOLOR protocol [6] (Fig. 1). WBV and regional volume are then calculated by multiplying the volume of a single voxel by the number of labeled voxels in original image space. In total, 15 NOIs are defined by structural and functional covariance networks including visual, frontal, language, memory, motor, fusiform, basal ganglia (BG) and cerebellum (CB).
2.2 Covariate-Adjusted Restricted Cubic Spline (C-RCS)
We define x as the ages of all subjects and \( S\left( x \right) \) as the corresponding brain volumes. In canonical nth degree spline regression, splines are used to model non-linear relationships between variables \( S\left( x \right) \) and x by deciding the connections between K knots \( (t_{1} < t_{2} < \cdots < t_{K} ) \). In this work, such knots were determined based on previously identified developmental shifts [1], specifically corresponding with transitions between childhood (7–12), late adolescence (12–19), young adulthood (19–30), middle adulthood (30–55), older adulthood (55–75), and late life (75–90). Using the expression from Durrleman and Simon [2], the canonical nth degree spline function is defined as
where \( \left( {x - t_{i} } \right)_{ + } = x - t_{i} , {\text{if }} x > t_{i} \); \( \left( {x - t_{i} } \right)_{ + } = 0, {\text{if }}x \le t_{i} \).
To regress out confound effects, new covariates \( X_{1}^{'} ,X_{2}^{'} , \ldots ,X_{c}^{'} \) (with coefficients \( \beta_{1}^{'} ,\beta_{2}^{'} , \ldots ,\beta_{c}^{'} \)) are introduced to the nth degree spline regression
where C is the number of confound effects.
In the RCS regression, a linear constrain is introduced [2] to address the poor behavior of the cubic spline model in the tails (\( x < t_{1} \;{\text{and}}\;x > t_{K} \)) [7]. Using the same principle, C-RCS regression extends the RCS regression (\( n = 3 \)) and restricts the relationship between \( S\left( x \right) \) and x to be a linear function in the tails. First, for \( x < t_{1} \),
where \( \dot{\beta }_{02} = \dot{\beta }_{03} = 0 \) ensures the linearity before the first knot. Second, for \( x > t_{K} \),
To guarantee the linearity of C-RCS after the last knot, we expand the previous expression and force the coefficients of \( x^{2} \) and \( x^{3} \) to be zero. After expansion,
As a result, linearity of \( S\left( x \right) \) at \( x > t_{K} \) implies that \( \sum\nolimits_{i = 1}^{K} {\dot{\beta }_{i3} } t_{i} = 0\; {\text{and}}\; \sum\nolimits_{i = 1}^{K} {\dot{\beta }_{i3} } = 0 \). Following such restrictions, the \( \dot{\beta }_{{\left( {K - 1} \right)3}} \) and \( \dot{\beta }_{K3} \) are derived as
and the complete C-RCS regression model is defined as
2.3 Regressing Out Confound Effects by C-RCS Regression in GLM Fashion
To adapt C-RCS regression in the GLM fashion, we redefine the coefficients \( \beta_{0} ,\;\beta_{1} ,\;\beta_{2} ,\; \ldots ,\;\beta_{K - 1} \) as Harrell [3] where \( \beta_{0} = \dot{\beta }_{00} ,\;\beta_{1} = \dot{\beta }_{01} ,\;\beta_{2} = \dot{\beta }_{13} ,\;\beta_{3} = \dot{\beta }_{23} ,\;\beta_{4} = \dot{\beta }_{33} ,\; \cdots ,\;\beta_{K - 1} = \dot{\beta }_{{\left( {K - 2} \right)3}} \). Then, the C-RCS regression with confound effects becomes
where C is the number for all confound effects (\( X_{u}^{'} \)). \( X_{1} = x \) and for \( j = 2, \ldots ,K - 1 \)
Then, the beta coefficients are solvable under GLM framework. Once \( \hat{\beta }_{0} ,\hat{\beta }_{1} , \hat{\beta }_{2} , \cdots ,\hat{\beta }_{K - 1} \) are obtained, two linear assured terms \( \hat{\beta }_{K} \) and \( \hat{\beta }_{K + 1} \) are estimated:
The final estimated volumetric trajectories \( \hat{S}(x) \) can be fitted as
In this work, gender, field strength and total intracranial volume (TICV) are employed as covariates \( X_{u}^{ '} \). TICV values are calculated using SIENAX [8]. Field strength and TICV are used to regress out site effects rather than using site categories directly since the sites are highly correlated with the explanatory variable age.
2.4 SCNs and CI Using Bootstrap Method
Using aforementioned C-RCS regression, the lifespan volumetric trajectories of WBV and 15 NOIs are obtained from 5111 images. Simultaneously, the piecewise volumetric trajectories within a particular age bin (between adjacent knots) of all 15 NOIs (\( \hat{S}_{i} \left( x \right), i = 1,2, \ldots ,15 \)) are separated to establish SCNs dendrograms using HCA [9]. The distance metric D used in HCA is defined as \( D = 1 - {\text{corr}}(\hat{S}_{i} \left( x \right),\;\hat{S}_{j} \left( x \right)),\;\;i,\;j \in \left[ {1,2, \ldots ,15} \right]\;{\text{and}}\;i \ne j \), where \( {\text{corr}}( \cdot ) \) is the Pearson’s correlation between any two C-RCS fitted piecewise trajectories \( \hat{S}_{i} \left( x \right) \) and \( \hat{S}_{j} \left( x \right) \) in the same age bin.
The stability of proposed approaches is demonstrated by the CIs of C-RCS regression and SCNs using bootstrap method [10]. First, the 95 % CIs of volumetric trajectories on WBV (Fig. 2) and 15 NOIs (Fig. 3) are derived by deploying C-RCS regression on 10,000 bootstrap samples. Then, the distances D between all pairs of clustered NOIs are derived using 15 (NOIs) × 10,000 (bootstrap) C-RCS fitted trajectories. Then, the 95 % CIs are obtained for each pair of clustered NOIs and shown on six SCNs dendrograms (Fig. 4). The average network distance (AND), the average distance between 15 NOIs for a dendrogram, can be calculated 10,000 times using bootstrap. The AND reflects the modularity of connections between all NOIs. We are able to see if the AND are significantly different during brain development periods by deploying the two-sample t-test on AND values (10,000/age bin) between age bins.
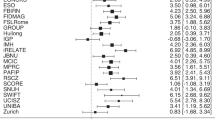
Volumetry and growth rate. The left plot in (a) shows the volumetric trajectory of whole brain volume (WBV) using C-RCS regression on 5111 MR images. The right figure in (a) indicates the growth rate curve, which shows volumetric change per year of the volumetric trajectory. In (b), C-RCS regression is deployed on the same dataset by additionally regressing out TICV. Our growth rate curves are compared with 40 previous longitudinal studies [1] on smaller cohorts (21 studies in (a) without regressing out TICV and 19 studies in (b) regressing out TICV). The standard deviations of previous studies are provided as black bars (if available). The 95 % CIs in all plots are calculated from 10,000 bootstrap samples.
Lifespan trajectories of 15 NOIs are provided with 95 % CI from 10,000 bootstrap samples. The upper 3D figures indicate the definition of NOIs (in red). The lower figures show the trajectories with CI using C-RCS regression method by regressing out gender, field strength and TICV (same model as Fig. 2b). For each NOI, the piecewise CIs of six age bins are shown in different colors. The piecewise volumetric trajectories and CIs are separated by 7 knots in the lifespan C-RCS regression rather than conducting independent fittings. The volumetric trajectories on both sides of each NOI are derived separately except for CB.
The six structural covariance networks (SCNs) dendrograms using hierarchical clustering analysis (HCA) indicate which NOIs develop together during different developmental periods (age bins). The distance on the x-axis is in log scale, which equals to one minus Pearson’s correlation between two curves. The correlation between NOIs becomes stronger from right to left on the x-axis. The horizontal range of each colored rectangles indicates the 95 % CI of distance from 10,000 bootstrap samples. Note that the colors are chosen for visualization purposes without quantitative meanings.
3 Results
Figure 2a shows the lifespan volumetric trajectories using C-RCS regression as well as the growth rate (volume change in percentage per year) of WBV when regressing out gender and field strength effects. Figure 2b indicates the C-RCS regression on the same dataset by adding TICV as an additional covariate. The cross sectional growth rate curve using C-RCS regression is compared with 40 previous longitudinal studies (19 are TICV corrected) [1], which are typically limited on smaller age ranges.
Using the same C-RCS model in Figs. 2b and 3 indicates the both lifespan and piecewise volumetric trajectories of 15 NOIs. In Fig. 4, the piecewise volumetric trajectories of the 15 NOIs within each age bin are clustered using HCA and shown in one SCNs dendrogram.
Then, six SCNs dendrograms are obtained by repeating HCA on different age bins, which demonstrate the evolution of SCNs during different developmental periods. The ANDs between any two age bins in Fig. 4 are statistically significant (p < 0.001).
4 Conclusion and Discussion
This paper proposes a large-scale cross-sectional framework to investigate life-time brain volumetry using C-RCS regression. C-RCS regression captures complex brain volumetric trajectories across the lifespan while regressing out confound effects in a GLM fashion. Hence, it can be used by researchers within a familiar context. The estimated volume trends are consistent with 40 previous smaller longitudinal studies. The stable estimation of volumetric trends for NOI (exhibited by narrow confidence bands) provides a basis for assessing patterns in brain changes through SCNs. Moreover, we demonstrate how to compute confidence intervals for SCNs and correlations between NOIs. The significant difference of AND indicates that the C-RCS regression detects the changes of average SCNs connections during the brain development.
The software is freely available onlineFootnote 1.
References
Hedman, A.M., van Haren, N.E., Schnack, H.G., Kahn, R.S., Hulshoff Pol, H.E.: Human brain changes across the life span: a review of 56 longitudinal magnetic resonance imaging studies. Hum. Brain Mapp. 33, 1987–2002 (2012)
Durrleman, S., Simon, R.: Flexible regression models with cubic splines. Stat. Med. 8, 551–561 (1989)
Harrell, F.: Regression Modeling Strategies: with Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis. Springer, Switzerland (2015)
Avants, B.B., Epstein, C.L., Grossman, M., Gee, J.C.: Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008)
Asman, A.J., Dagley, A.S., Landman, B.A.: Statistical label fusion with hierarchical performance models. In: Proceedings - Society of Photo-Optical Instrumentation Engineers, vol. 9034, p. 90341E (2014)
Klein, A., Dal Canton, T., Ghosh, S.S., Landman, B., Lee, J., Worth, A.: Open labels: online feedback for a public resource of manually labeled brain images. In: 16th Annual Meeting for the Organization of Human Brain Mapping (2010)
Stone, C.J., Koo, C.-Y.: Additive splines in statistics, p. 48 (1986)
Smith, S.M., Zhang, Y., Jenkinson, M., Chen, J., Matthews, P.M., Federico, A., De Stefano, N.: Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage 17, 479–489 (2002)
Anderberg, M.R.: Cluster Analysis for Applications: Probability and Mathematical Statistics: A Series of Monographs and Textbooks. Academic Press, New York (2014)
Efron, B., Tibshirani, R.J.: An Introduction to the Bootstrap. CRC Press, Boca Raton (1994)
Acknowledgments
This research was supported by NSF CAREER 1452485, NIH 5R21EY024036, NIH 1R21NS064534, NIH 2R01EB006136, NIH 1R03EB012461, NIH R01NS095291 and also supported by the Intramural Research Program, National Institute on Aging, NIH.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Huo, Y., Aboud, K., Kang, H., Cutting, L.E., Landman, B.A. (2016). Mapping Lifetime Brain Volumetry with Covariate-Adjusted Restricted Cubic Spline Regression from Cross-Sectional Multi-site MRI. In: Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9900. Springer, Cham. https://doi.org/10.1007/978-3-319-46720-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-46720-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46719-1
Online ISBN: 978-3-319-46720-7
eBook Packages: Computer ScienceComputer Science (R0)