Abstract
Understanding patterns of brain development before birth is of both high clinical and scientific interest. However, despite advances in reconstruction methods, the challenging setting of in-utero imaging renders precise, point-wise measurements of the rapidly changing fetal brain morphology difficult. This paper proposes a method to deal with bad measurement quality due to image noise, motion artefacts and ensuing segmentation and registration errors by enforcing spatial regularity during the estimation of parametric models of cortical expansion. Qualitative and quantitative analysis of the proposed method was performed on 88 clinical fetal MR volumes. We show that the resulting models accurately capture the morphological and temporal properties of fetal brain development by predicting gestational age on unseen cases at human-level accuracy.
G. Langs—This project was supported by the FWF under KLI 544-B27 and I2714-B31, the OeNB under 14812 and 15929, and EU FP7 under 2012-PIEF-GA-33003.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
During the second and third trimester of gestation, the fetal brain grows from a smooth shape to a complex folded structure. Understanding the processes driving this rapid development is of strong academic and clinical interest [1]. In the last decade, in utero imaging using Magnetic Resonance Imaging (fetal MRI), together with specialized reconstruction procedures [2] have led to a better understanding of gross morphological fetal neurodevelopment. Various authors have reported normative values for the developing brains’ volume, folding and surface area. However, these studies are either based on premature neonates [3], report global measurements [4–6] or rely on a-priori parcellations of the cortical surface into lobar regions [7].
In this paper, we aim at computing a continuous model of fetal cortical expansion. We build on recent advances in structured prediction [8] to regularize parametric growth models along the cortex. We show that the resulting models can be used to precisely model fetal cortical expansion on a surface node level, predict gestational age with high accuracy, and identify cortical regions that are predictive for age.
2 Regularizing Parametric Cortical Growth Models
We propose a method allowing for the joint estimation of sets of parametric models in a regularization framework. Modeling prior knowledge about the type of interactions between elements of the set as graphs allows us to include this information in the process of fitting the models to noisy data. Specifically, we are interested in enforcing spatial smoothness of growth models on a surface. We show how this can be achieved in the case of a special type of logistic growth model, the Gompertz function.
Parametric Growth Modeling. Simple models such as linear and polynomial functions are commonly used to summarize observations. However, their underlying assumption that the observed process is unbounded is generally not valid. Logistic functions such as the Gompertz function
on the other hand can be used to describe an asymptotically bounded processes such as brain growth [7]. Fitting functions of this type to measurements allows for the summarization of the observed process in terms of initial (\(\beta _1\)) and final (\(\beta _2\)) quantity, as well as the timing (\(\beta _4\)) and rate (\(\beta _3\)) of its growth.
Graph-Based Regularization. Spatial relationships between observations can be incorporated into model fitting using an appropriate regularization term. This approach is common in imaging, where the lattice structure of image elements is exploited to solve otherwise ill-posed problems such as denoising, segmentation or registration. In linear modeling, more complex types of structured approaches have been proposed that exploit known regularities of the data such as sparsity or group structure. In work that is most related to the proposed method, Grosenik et al. [8] exploit the underlying spatial coherence of fMRI to predict subjects’ behavior by solving the regularized linear least squares problem
where G is a graph representing the connections between voxels of the fMRI volume.
Spatially Smooth Gompertz Models. A common measure of smoothness of a function \(f: \mathbb {R}^n \rightarrow \mathbb {R}^m\) is the sum of all its unmixed second partial derivatives, which define the Laplace operator \(\varDelta f = \sum _{i=1}^n \frac{\partial ^2 f}{\partial x_i^2}\). In the discrete space of values observed on a graph G, an analoguous operator can be defined as \(L = D - A\), where D is the diagonal matrix of the degrees of the nodes in the adjacency matrix A.
We introduce this prior into the non-linear problem of fitting a set of growth models represented as Gompertz functions to observations obtained at locations corresponding to nodes in a graph G. As in [8], we encourage spatial smoothness of the model parameters in G by solving
where \(\varvec{\beta } = (\beta _{(1,1)},\dots ,\beta _{(4,1)},\dots ,\beta _{(1,n)},\dots ,\beta _{(4,n)})\) and \(y(t,\mathbf {x}_n)\) is a measurement at location \(\mathbf {x}_n\) at time t. Introducing the diagonal matrix \(B \in \mathbb {R}^{4\times 4}\) allows us to selectively penalize spatial variablity of specific model parameters. For example, setting \(B = {\text {diag}}\begin{pmatrix}0,&0,&1,&1\end{pmatrix}\) adds costs for the rate and timing of growth, but not its amplitude. We solve Eq. 3 for observed data y using constrained optimization to ensure \(\beta _{(3,\cdot )} > 0\).
3 Data, Cortical Segmentation, and Tracking
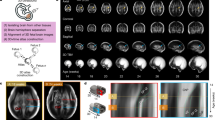
Obtaining surface models of the fetal brain from fetal MRI requires a sequence of processing steps. Artefacts due to fetal motion during the acquisition are mitigated by using fast Rapid Acquisition with Refocused Echoes (RARE) T2 sequences [9] at increased (3–4 mm) slice thickness. To avoid the loss of important anatomical information due to this strong anisotropy, orthogonal views in axial, coronal and sagittal direction are acquired and fused into an isotropic, high-resolution (HR) volume [2].
Cortical Segmentation. We use probability maps of brain tissues provided with a publicly available atlas of fetal brain anatomy [10] to initialize a graph-based segmentation procedure [11]. Atlas alignment is performed using affine and non-rigid registration, initialized using fiducials placed at the distal horns of the lateral ventricles. The resulting segmentation is split at the interhemispheric fissure and an estimate of the gray/white matter boundary in each hemisphere is computed using marching cubes. A spherical parametrization of the resulting surface mesh is then computed [12] and used to obtain an initial regular sampling \(S_0=(V_0, F_0)_{\{l,r\}}, |V_0|=642, |F_0|=1280\) of each cortical surface. The surface mesh of each hemisphere is deformed towards the darkest part of the cortical band using an active contour model while avoiding self-intersections. We proceed in a multi-resolution manner using recursive icosahedral subdivisions. This procedure stops after 3 subdivisions, yielding surface meshes \(S_3=(V_3,F_3)_{\{l,r\}}, |V_3|=40962, |F_3|=81920\).
Surface Normalization. Comparing the surfaces of different cases requires their point-wise correspondence. We employ a spectral graph matching technique [13] to establish inter-patient correspondences. To ensure compatibility over the whole course of development, we first perform surface extraction on the atlas volumes of [10] and compute pair-wise correspondences between the models obtained for subsequent weeks as described in [14]. We then perform spectral matching between each individual case and its age-matched reference surface. After normalization, we compute the surface area of each face in \(F_3\) using Heron’s formula. As \(F_{n+1}\) is a complete subdivision of \(F_n\), we can directly integrate these measurements over the faces of \(F_0\) to reduce computational complexity. Note that this is not identical to computing the area of \(F_0\) directly.
4 Results
We performed the described reconstruction, segmentation and matching procedure on 88 fetal MRIs obtained from routine clinical evaluations at the General Hospital of Vienna. The gestational age in days (GW + d, or GD) of the individual cases has been assessed based on the last menstrual cycle of the mother. The dataset spans the whole second trimester from GW 19 + 5 to GW 36 + 5 (mean GD \(201.76\pm 35.34\)). None of the cases showed any neurological pathologies.
A Continuous Model of Fetal Cortical Expansion. We fit both independent and graph-regularizedFootnote 1 Gompertz models to measurements of local surface area of corresponding faces in the 88 surface models. Due to differences in the size of the surface elements in the triangulation (Fig. 1(b)), we enforce smoothness only on the growth rate \(\beta _{(3,\cdot )}\) and inflation time-point \(\beta _{(4,\cdot )}\) by setting B in Eq. 3 accordingly.
Inter-patient variability as well as inconsistencies in the segmentation and matching procedures lead to considerable noise when measuring the local surface area (Fig. 1). Thus, fitting independent growth models at each location \(\mathbf {x}\) can yield physiologically meaningless measurements. This is clearly visible in Fig. 1(a), where the time-courses of development estimated via the fitted models vary considerably in neighboring surface patches. Using the proposed graph-regularized model on the other hand (Fig. 1(c)) has the expected effect of finding locally similar models that still fit the data well. We assesed model fit by computing \(R^2\) values for both the individual (\(R^2=0.75\)) and regularized (\(R^2=0.74\)) model, which yielded only a negligeable (95 % CI \([9\cdot 10^{-3}, 1\cdot 10^{-2}]\)) decrease for the regularized model. Lower model fit could be expected due to the added regularization term in Eq. 3. The small difference however shows that the proposed hypothesis of a smooth developmental process of fetal cortical expansion can indeed be experimentally validated.
A major advantage of logistic models are their interpretable parameters. Regularization plays an important role of providing more robust measures of these factors by reducing the influence of spurious measurements on the model fit. Figures 2–4 show this effect when modeling fetal cortical expansion. The relative increase in cortical surface area (Fig. 2) corresponds well to published results on the global cortical expansion [5, 6] and due to our choice of B remains largely unaffected by the regularization. On the other hand, both the time-point of maximal cortical expansion (Fig. 3) and the rate of growth (Fig. 4) show the expected effect of the regularization removing physiologically implausible results such as an inhomogenous time-course of operculization or an inhomogeneous expansion pattern in the insula.
Note however that the regularized model fit does not simply correspond to smoothing the parameters of the unregularized model on the surface mesh. This is apparent in the absence of the distinctive blurring effect of high values affecting its neighbors, as for example in the expansion rate at the level of the right middle frontal gyrus (Fig. 4) or in the expansion time-point of the right superior frontal sulcus (Fig. 3).
Predicting Gestational Age from Cortical Surface Area. Given a measurement of local cortical surface area and a fitted model \(\beta _{1\ldots 4}\), the inverse of the Gompertz model can be used to estimate the gestational age of a fetusFootnote 2. We evaluate the validity of models obtained with \(\lambda \in [0, 0.1, 0.5, 1, 2, 5, 10]\) using Leave-One-Out Cross-Validation (LOOCV). By averaging all predictions of the models, we achieved an overall error of \(5.70\pm 4.49\) days in the unregularized case. The best results using regularized models were obtained using \(\lambda = 5\), yielding a prediction error of \(5.70\pm 4.37\) days. Although these differences are not significant, the regularized model is much easier to interpret. This shows that spatial smoothness is an adequate prior for fetal cortical expansion and helps in building descriptive, interpretable growth models. Prediction accuracy in both cases is also much higher as for instance in [7] where the authors used global cortical surface curvature measurements instead of local ones. We plot the GWs estimated using the regularized model against the true GWs in Fig. 5(a), along with their 0.95-th quantiles over all model faces. Comparing the error rates for the left and right hemispheres shows an increase in variability with age that is higher in the left than the right hemisphere.
Finally, the proposed method allows for the point-wise evaluation of the prediction error (Fig. 5(b)). This enables us to distinguish regions that are stable predictors of age, such as the parietal lobe, the prefrontal cortex and the callosal area. We performed an exhaustive search over percentiles of face-wise LOOCV prediction errors in the range of \([0.01,\ldots , 1]\) to determine a threshold defining regions predictive of gestational age. The best prediction was obtained by retaining all faces that have a mean prediction error below 11.79 days (GW 1 + 5, 0.05-th quantile for \(\lambda =5\)), Fig. 5(b). By averaging the prediction in these regions, the error is reduced significantly to \(4.65\pm 3.58\) days (\(p<0.05\)). The accuracy of these results is in the order of the underlying uncertainty of reported last menstrual cycle and comparable with state of the art results in [15] as well as manual measurements [4].
5 Conclusion
We have proposed a novel method for fitting spatially regularized growth models to noisy data. Applying this method in the challenging setting of fetal brain development enables building accurate interpretable models of cortical expansion in utero, and allows for the point-wise estimation of gestation age. We have shown that the resulting models are in line with published knowledge about fetal brain growth and are able to predict the age of the fetus with high accuracy. We believe that the presented method is of significant value in deepening the understanding of the time-course of neuroanatomical development, as well as allowing for the precise localization and characterization of its vulnerabilities.
Notes
- 1.
The choice of the regularization weight \(\lambda \) is described in the next section.
- 2.
The inverse of Eq. 1 is given as \(t = f^{-1}(y) = -\frac{\log (-\log ((y-\beta _1)/\beta _2))}{\beta _3} + \beta _4\).
References
Tallinen, T., Chung, J.Y., Rousseau, F., Girard, N., Lefevre, J., Mahadevan, L.: On the growth and form of cortical convolutions. Nat. Phys. Feburary 2016
Rousseau, F., Oubel, E., Pontabry, J., Schweitzer, M., Studholme, C., Koob, M., Dietemann, J.L.: BTK: An open-source toolkit for fetal brain MR image processing. Comput. Methods Prog. Biomed. 191(1) (2012)
Dubois, J., Benders, M., Cachia, A., Lazeyras, F., Leuchter, R.H.V., Sizonenko, S.V., Borradori-Tolsa, C., Mangin, J.F., Hüppi, P.S.: Mapping the early cortical folding process in the preterm newborn brain. Cereb. Cortex 18(6), 1444–1454 (2008)
Wu, J., Awate, S.P., Licht, D.J., Clouchoux, C., du Plessis, A.J., Avants, B.B., Vossough, A., Gee, J.C., Limperopoulos, C.: Assessment of MRI-based automated fetal cerebral cortical folding measures in prediction of gestational age in the third trimester. Am. J. Neuroradiol. 36(7), 1369–1374 (2015)
Clouchoux, C., Kudelski, D., Gholipour, A., Warfield, S.K., Viseur, S., Bouyssi-Kobar, M., Mari, J.L., Evans, A.C., du Plessis, A.J., Limperopoulos, C.: Quantitative in vivo MRI measurement of cortical development in the fetus. Brain Struct. Funct. 217(1), 127–139 (2011)
Rajagopalan, V., Scott, J., Habas, P.A., Kim, K., Corbett-Detig, J., Rousseau, F., Barkovich, A.J., Glenn, O.A., Studholme, C.: Local tissue growth patterns underlying normal fetal human brain gyrification quantified in utero. J. Neurosci. 31(8), 2878–2887 (2011)
Wright, R., Kyriakopoulou, V., Ledig, C., Rutherford, M.A., Hajnal, J.V., Rueckert, D., Aljabar, P.: Automatic quantification of normal cortical folding patterns from foetal brain MRI. NeuroImage 91, 1–12 (2014)
Grosenick, L., Klingenberg, B., Katovich, K., Knutson, B., Taylor, J.E.: Interpretable whole-brain prediction analysis with GraphNet. NeuroImage 72(C), 304–321 (2013)
Hennig, J., Nauerth, A., Friedburg, H.: RARE imaging: a fast imaging method for clinical MR. Magn. Reson. Med. 3(6), 823–833 (1986)
Serag, A., Aljabar, P., Ball, G., Counsell, S.J., Boardman, J.P., Rutherford, M.A., Edwards, A.D., Hajnal, J.V., Rueckert, D.: Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. NeuroImage 59(3), 2255–2265 (2012)
Rajchl, M., Baxter, J.S.H., McLeod, A.J., Yuan, J., Qiu, W., Peters, T.M., Khan, A.R.: Hierarchical max-flow segmentation framework for multi-atlas segmentation with Kohonen self-organizing map based Gaussian mixture modeling. Med. Image Anal. 1–19, May 2015
Crane, K., Pinkall, U., Schröder, P.: Robust fairing via conformal curvature flow. ACM Trans. Graph. 32(4), 1–10 (2013)
Lombaert, H., Sporring, J., Siddiqi, K.: Diffeomorphic spectral matching of cortical surfaces. In: Gee, J.C., Joshi, S., Pohl, K.M., Wells, W.M., Zöllei, L. (eds.) IPMI 2013. LNCS, vol. 7917, pp. 376–389. Springer, Heidelberg (2013). doi:10.1007/978-3-642-38868-2_32
Wright, R., Makropoulos, A., Kyriakopoulou, V., Patkee, P.A., Koch, L.M., Rutherford, M.A., Hajnal, J.V., Rueckert, D., Aljabar, P.: Construction of a fetal spatio-temporal cortical surface atlas from in utero MRI: application of spectral surface matching. NeuroImage 120(C), 467–480 (2015)
Namburete, A.I.L., Stebbing, R.V., Kemp, B., Yaqub, M., Papageorghiou, A.T., Noble, J.A.: Learning-based prediction of gestational age from ultrasound images of the fetal brain. Med. Image Anal. 21(1), 72–86 (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Schwartz, E., Kasprian, G., Jakab, A., Prayer, D., Schöpf, V., Langs, G. (2016). Modeling Fetal Cortical Expansion Using Graph-Regularized Gompertz Models. In: Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9900. Springer, Cham. https://doi.org/10.1007/978-3-319-46720-7_29
Download citation
DOI: https://doi.org/10.1007/978-3-319-46720-7_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46719-1
Online ISBN: 978-3-319-46720-7
eBook Packages: Computer ScienceComputer Science (R0)