Abstract
Investigating functional brain networks and activities using sparse representation of fMRI data has received significant interests in the neuroimaging field. It has been reported that sparse representation is effective in reconstructing concurrent and interactive functional brain networks. However, previous data-driven reconstruction approaches rarely simultaneously take consideration of anatomical structures, which are the substrate of brain function. Furthermore, it has been rarely explored whether structured sparse representation with anatomical guidance could facilitate functional networks reconstruction. To address this problem, in this paper, we propose to reconstruct brain networks using the anatomy-guided structured multi-task regression (AGSMR) in which 116 anatomical regions from the AAL template as prior knowledge are employed to guide the network reconstruction. Using the publicly available Human Connectome Project (HCP) Q1 dataset as a test bed, our method demonstrated that anatomical guided structure sparse representation is effective in reconstructing concurrent functional brain networks.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Functional magnetic resonance imaging (fMRI) signal analysis and functional brain network investigation using sparse representation has received increasing interests in the neuroimaging field [1, 10]. The main theoretical assumption is that each brain fMRI signal can be represented as sparse linear combination of a set of signal basis in an over-complete dictionary. The data-driven strategy of dictionary learning and sparse coding is efficient and effective in reconstructing concurrent and interactive functional networks from both resting state fMRI (rsfMRI) and task base fMRI (tfMRI) data [1, 10]. However, these approaches have potential space of further improvement, because the pure data-driven sparse coding does not integrate brain science domain knowledge when reconstructing functional networks. In the neuroscience field, it is widely believed that brain anatomy and structure play crucial roles in determining brain function, and anatomical structure is the substrate of brain function. Thus integrating anatomical structure information into brain network representation is well motivated and justified.
In this paper, we propose a novel anatomy-guided structured multi-task regression (AGSMR) method for functional network reconstruction by employing anatomical group structures to guide sparse representation of fMRI data. In general, group-wise structured multi-task regression has been an established methodology, which puts group structure on the multi-tasks and employs a combination of \( \ell_{2} \) and \( \ell_{1} \) norms in order to learn both intra-group homogeneity and inter-group sparsity [2, 6]. Our premise is that fMRI voxels from the same anatomical structure should potentially play similar role in brain function. Thus, employing 116 brain regions from the AAL template as anatomical group information could effectively improve the network representation by constraining both homogeneity within anatomical structure and sparsity across anatomical structures. After applying our method on the recently publicly released Human Connectome Project (HCP) data, our experimental results demonstrate that networks have been improved with higher similarity, which also provides anatomical clues for understanding the detected brain networks.
2 Method
2.1 Overview
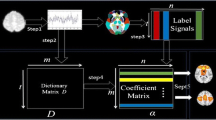
Our computational framework of AGSMR is illustrated in Fig. 1. fMRI images from individual brain are first registered into a standard space(MNI) to align with the AAL template. Then extracting fMRI signals from a whole brain mask, an over-complete signal dictionary is learned via online dictionary method. The learned dictionary as a set of features (regressors), the group structured multi-task regression employs anatomical structures as group information to regress whole brain signals. Finally, the coefficients matrix are mapped back to the brain volume represent functional brain networks.
The flowchart of proposed AGSMR method pipeline: Step 1: data acquisition, preprocessing and extract the whole brain signals. Step 2: using the whole signals for learning dictionary D. Step 3: labelling of the whole signals via the AAL template. Step 4: feature Selection based on AGSMR method. Step 5: mapping the selected feature (coefficient matrix) in the whole brain to identify these meaningful functional networks.
2.2 Data Acquisition and Preprocessing
The recently publicly released fMRI data by Human Connectome Project (HCP) (Q1) was used in this paper. The dataset (Q1) was acquired for 68 subjects and it includes 7 tasks such as Motor, Emotion, Gambling, Language, Relational, Social, and Working Memory. The acquisition parameters of tfMRI data are as follows: 90 × 104 matrix, 220 mm FOV, 72 slices, TR = 0.72 s, TE = 33.1 ms, flip angle = 52°, BW = 2290 Hz/Px, in-plane FOV = 208 × 180 mm, 2.0 mm isotropic voxels. The preprocessing pipelines included motion correction, spatial smoothing, temporal pre-whitening, slice time correction, global drift removal. More details about the task descriptions and preprocessing are referred to [9]. After preprocessing, all fMRI images are registered into a standard template space (MNI space). Then fMRI signals are extracted from voxels within a brain mask, and each signal was normalized to be with zero mean and standard deviation of 1.
2.3 The Whole Brain Signals Dictionary Learning
In our method, an over-complete dictionary \( D \) is first learned from the whole brain fMRI signals \( X = [x_{1} ,x_{2} , \ldots x_{n} ] \in R^{t \times n} \) (t is the fMRI signal time point and n is the voxel number) using online dictionary learning method [4]. The theoretical assumption here is that the whole brain fMRI signals are represented by sparse linear combination of a set of signal basis, i.e., dictionary atoms. The empirical cost function of learning is defined in Eq. (1)
where \( D = [d_{1} ,d_{2} , \ldots d_{n} ] \in R^{t \times m} \)(t is the fMRI signal time point and m is the number of dictionary atoms) is the dictionary, each column representing a basis vector, the \( \ell_{1} \) regularization in Eq. (2) was adopted to generate a sparse solution, \( D \) and \( \alpha \) are alternatively updated and learned by using online dictionary learning algorithm [4]. The learned \( D \) was adopted as the features (regressors) to perform sparse representation and the proposed structured sparse representation of brain fMRI signals is detailed in Sect. 2.5.
2.4 Grouping fMRI Signals with Anatomical AAL Template
By using the AAL template [7], 116 brain regions are employed in our method as shown in Fig. 1. Specially, the whole brain voxel are separated into 116 groups based on AAL template. Before signal extraction, each subject has been registered into the standard space(MNI) and alignment is established with the AAL template, where each voxel in brain mask is associated with a template label. Voxels with same anatomical AAL label are grouped together. Thus, in each brain, voxels of fMRI signals are categorized and labeled as 116 AAL groups. This anatomical group information will be used to guide the coefficient matrix learning in the next section.
2.5 Anatomical Guided Structured Multi-task Regression (AGSMR)
In conventional approach, once the dictionary \( D \) are defined, the learning of coefficient matrix is summarized into the typical LASSO [5] problem in Eq. (3).
where \( \ell (\alpha ) \) is the loss function, and \( \phi (\alpha ) \) is the regularization term, which could regularize feature selection while achieving sparse regularization, and λ > 0 is the regularization parameter. Once we learned dictionary \( D = [d_{1} ,d_{2} , \ldots d_{n} ] \in R^{t \times m} \) (Sect. 2.3), the conventional LASSO perform regression of brain fMRI signals \( X = [x_{1} ,x_{2} , \ldots x_{n} ] \in R^{t \times n} \) to obtain a sparse coefficient matrix \( \alpha = [\alpha_{1} ,\alpha_{2} , \ldots \alpha_{n} ] \in R^{m \times n} \) was defined as:
where \( \ell (\alpha ) \) is defined as the least square loss, and \( \phi (\alpha ) \) is the \( \ell_{1} \)-norm regularization term to induce sparsity, \( \alpha_{i}^{j} \) is the coefficient element at the i-th column and j-th row, \( m \) is the dictionary size. Equation (4) can be viewed as the LASSO penalized least squares problem, conventional LASSO in Eq. (4) is pure data-driven approach, However, according to the previous studies [2, 3, 6] that have shown that the priori structure information such as disjoint/overlapping groups, trees, and graphs may significantly improve the classification/regression performance and help identify the important features [3].
In this paper, we propose a novel structured sparse representation approach (group guided structured multi-task regression) into the regression of fMRI signals. Specifically, the group information of fMRI signals are defined by the anatomical structure in Sect. 2.4, i.e., the whole brain fMRI signals are separated into \( v \) groups \( \{ G_{1} ,G_{2} , \ldots G_{v} \} ,v = 1,2, \ldots V \) based on the AAL template. The conventional LASSO adopted the \( \ell_{1} \) norm regularization term to induce sparsity (Eq. (4)), here the \( \ell_{2} \) norm penalty is introduced into the penalty term as shown in Eq. (5), which will improve the intra-group homogeneity. Meanwhile, we using \( \ell_{1} \) norm joint \( \ell_{2} \) norms penalty which will induce both intra-group sparsity and inter-group sparsity in Eq. (5).
Thus, Eq. (5) can be also viewed as the structured sparse penalized multi-task least squares problem. The detailed solution of this structured LASSO penalized multi-task least squares problem with combined \( \ell_{1} \) and \( \ell_{2} \) norms were referred to [6, 8] our final learning problem is summarized in Eq. (5). http://yelab.net/software/SLEP/) is the SLEP package employed to solve the problem and to learn the coefficient matrix \( \alpha \). From brain science perspective, the learned coefficient matrix \( \alpha \) include the spatial feature of functional networks and each row of \( \alpha \) spatial features were mapped back to brain volume to identify and quantitatively characterize those meaningful functional networks similar to the methods in [1].
3 Results
3.1 Identifying Resting State Networks on Seven Task Datasets
To evaluate the identified networks, we defined a spatial similarity coefficient for check the spatial similarity between the identified networks and the resting state networks (RSNs) template [11]. The similarity coefficient was defined as below:
where A is the spatial map of our identified network component and B is that of the RSNs template network. \( |A| \) And \( |B| \) are the numbers of voxels.
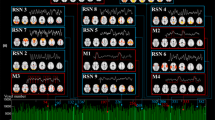
We performed quantitative measurements on Working Memory task dataset to demonstrate the performance of our method. We selected 10 well-known resting state networks to compare spatial similarity. The identified networks are visualized in Fig. 2. The figures(RSNs#1—RSNs#10) represent 10 resting state template networks(RSNs) and #1–#10 represent our identified networks. It is shown that our method identified networks are consistent with the templates. The slice #1, #2, and #3 are visual network, which correspond to medial, occipital pole, and lateral visual areas, the slice #4 is default mode network(DMN),the slice #5 to #8 are cerebellum, sensorimotor, auditory and executive control networks respectively. The slice #9 and #10 are frontoparietal networks, all of these identified networks activated areas are consistent with template networks and the detailed comparision results in Table 1.
Comparison 10 resting state networks (RSNs) with our method identified networks on working memory task dataset. The figures(RSNs#—1RSNs#10) show 10 resting state template networks [11] and (#1–#10) our method identified networks.
In order to validate our method effective and robust, we used seven different task datasets to test our approach. Figure 3 shows the results. Table 1 shows similarity results compare with template on 7 different datasets.
3.2 Comparison Between Our Method and Traditional Method
In this section, we compare our method and LASSO method on both Working Memory and Gambling datasets. Figure 4 shows the our method and LASSO method identified visual network, executive control network and auditory network, respectively. Table 2 shows the two methods similarities comparisons results with the template on two different task datasets. These comparisons show that our method has higher similarity with the template, and in this sense it is superior in reconstructing functional networks than no used anatomical structure the traditional method of LASSO.
4 Conclusion
In this paper, we propose a novel anatomy guided structured multi-task regression method for brain network identification. Experiments based on 7 different task datasets have demonstrated the effectiveness of our AGSMR method in identifying consistent brain networks. Comparisons have shown that our method is more effective and accurate than the traditional method of LASSO. In general, our approach provides the anatomical substrates for the reconstructed functional networks. In the future, we plan to apply and test this AGSMR method in larger fMRI datasets and compare it with other brain network construction methods. In addition, it will be applied on clinical fMRI datasets to potentially reveal the abnormalities of brain networks in diseased brains.
References
Lv, J., Jiang, X., Li, X., Zhu, D., Chen, H., Zhang, T., Hu, X., Han, J., Huang, H., Zhang, J.: Sparse representation of whole-brain fMRI signals for identification of functional networks. Med. Image Anal. 20, 112–134 (2015)
Kim, S., Xing, E.P.: Tree-guided group lasso for multi-task regression with structured sparsity. In: ICML, pp. 543–550 (2010)
Ye, J., Liu, J.: Sparse methods for biomedical data. ACM SIGKDD Explor. Newslett. 14, 4–15 (2012)
Mairal, J., Bach, F., Ponce, J., Sapiro, G.: Online learning for matrix factorization and sparse coding. J. Mach. Learn. Res. 11, 19–60 (2010)
Tibshirani, R.: Regression shrinkage and selection via the lasso. J. Roy. Stat. Soc. Ser. B (Methodological) 58, 267–288 (1996)
Liu, J., Ji, S., Ye, J.: Multi-task feature learning via efficient l 2, 1-norm minimization. In: Proceedings of the Twenty-Fifth Conference on Uncertainty in Artificial Intelligence, pp. 339–348. AUAI Press (2009)
Tzourio-Mazoyer, N., Landeau, B., Papathanassiou, D., Crivello, F., Etard, O., Delcroix, N., Mazoyer, B., Joliot, M.: Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289 (2002)
Liu, J., Ji, S., Ye, J.: SLEP: Sparse learning with efficient projections. Arizona State University (2009)
Van Essen, D.C., Smith, S.M., Barch, D.M., Behrens, T.E., Yacoub, E., Ugurbil, K.: WU-Minn HCP consortium. The WU-Minn human connectome project: an overview. Neuroimage 80, 62–79 (2013)
Lv, J., Jiang, X., Li, X., Zhu, D., Zhang, S., Zhao, S., Chen, H., Zhang, T., Hu, X., Han, J., Ye, J.: Holistic atlases of functional networks and interactions reveal reciprocal organizational architecture of cortical function. IEEE Trans. Biomed. Eng. 62, 1120–1131 (2015)
Smith, S., Fox, P., Miller, K., Glahn, D., Fox, P., Mackay, C., Filippini, N., Watkins, K., Toro, R., Laird, A., Beckmann, C.: Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. U.S.A. 106, 13040–13045 (2009)
Acknowledgements
This research was supported in part by Jiangsu Natural Science Foundation (Project No. BK20131351), by the Chinese scholarship council (CSC).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Zhao, Q., Lu, J., Lv, J., Jiang, X., Zhao, S., Liu, T. (2016). Exploring Brain Networks via Structured Sparse Representation of fMRI Data. In: Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9900. Springer, Cham. https://doi.org/10.1007/978-3-319-46720-7_7
Download citation
DOI: https://doi.org/10.1007/978-3-319-46720-7_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46719-1
Online ISBN: 978-3-319-46720-7
eBook Packages: Computer ScienceComputer Science (R0)