Abstract
Several neurological disorders are associated with hippocampal pathology. As changes may be localized to specific subfields or spanning across different subfields, accurate subfield segmentation may improve non-invasive diagnostics. We propose an automated subfield segmentation procedure, which combines surface-based processing with a patch-based template library and feature matching. Validation experiments in 25 healthy individuals showed high segmentation accuracy (Dice >82 % across all subfields) and robustness to variations in the template library size. Applying the algorithm to a cohort of patients with temporal lobe epilepsy and hippocampal sclerosis, we correctly lateralized the seizure focus in >90 %. This advantageously compares to classifiers relying on volumes retrieved from other state-of-the-art algorithms.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The hippocampus plays a key role in cognition and its compromise is a hallmark of several prevalent brain disorders, such as temporal lobe epilepsy (TLE) [1]. With the advent of large-scale neuroimaging data basing and analysis in health and disease, the development of accurate automated segmentation approaches becomes increasingly important.
The majority of automated hippocampal segmentation approaches have operated on a global scale. Recent methods rely on a multi-template framework to account for interindividual anatomical variability. While the majority of previous algorithms employed a purely voxel-based strategy, adopting a surface-based library has shown benefits by improving flexibility to model shape deformations often seen in disease, but also in 10–15 % of healthy subjects [2]. To improve label fusion and image matching, recent studies have adopted patch-based methods that compactly represent shape, anatomy, texture, and intensity [3]. Notably, these approaches have also been successful in non-segmentation tasks, such as image denoising [4] and supersampling [5].
Developments in MRI hardware have begun to generate images of brain anatomy with unprecedented details [6], fostering guidelines to manually delineate hippocampal subfields. Few automated methods have been proposed, each relying on multiple templates together with: (i) Bayesian inference and fusion of ex-vivo/in-vivo landmarks [7, 8], (ii) label propagation to intermediate templates [9], (iii) combinations of label fusion (taking inter-template similarity into account) and post-hoc segmentation correction [10]. These methods operate either on anisotropic T2-weighted images [10], or T1-weighted images with standard millimetric [8, 9], or submillimetric resolution [7]. Only one study so far [9] calculated Dice overlaps of manual and automated labels in millimetric T1-weighted images, with modest performance (Dice: 0.56–0.65).
We propose a novel approach for hippocampal subfield segmentation SurfPatch, which combines multi-template feature matching with deformable parametric surfaces and vertex-wise patch sampling, relying on point-wise correspondence across the template library. Validation was performed using a publically available 3T dataset of manual segmentations together with high- and standard-resolution MRI data of healthy controls [11]. We also applied SurfPatch to 17 TLE patients with hippocampal atrophy, testing its ability to lateralize the seizure focus and compared its performance to two ASHS (Automatic Segmentation of Hippocampal Subfields) [10] and FreeSurfer 5.3 [7].
2 Methodology
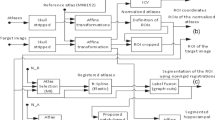
Figure 1 below summarizes the segmentation steps.
For training, SurfPatch builds the mean patch surface \( S^{\mu } \) and standard deviation (SD) patch surface \( S^{\sigma } \) across the template library (Fig. 1A). For segmentation, it nonlinearly warps each template surface to the test case, re-computes patch features across the warped surface, and normalizes features using surface-based z-scoring (relative to \( S^{\mu } \) and \( S^{\sigma } \)). Based on vertex-wise z-score, it selects a subset of templates, builds an average surface, and performs a deformation for final segmentation (Fig. 1B).
2.1 Training Step (Fig. 1A)
Subfield labels are converted to surface meshes and parameterized using spherical harmonics and a point distribution model (SPHARM-PDM) [12] that guarantees correspondence of surface points (henceforth, vertices) across subjects (Fig. 1 A-1). For a given template t, its reconstructed surface \( S^{t} \) is mapped to its corresponding T1-MRI \( I^{t} \). Let \( x_{k}^{v} \) with \( k \in \left\{ {1, \ldots ,8} \right\} \) be the eight closest voxels of a given vertex \( v \), and let \( P_{x,k}^{t} \) be the corresponding local cubical neighborhoods (i.e., patches) centered around these voxels. We build a vertex patch \( P_{v}^{t} \) by computing a trilinear interpolation of these 8 patches (which is an extension of the trilinear interpolation of the 8 closest voxels). Patches are considered as vectors. By pooling corresponding vertex patches from each template surface, we derive the mean \( P_{v}^{\mu } \) and SD patch \( P_{v}^{\sigma } \) at vertex \( v \):
where N is the number of templates (Fig. 1A-2).
2.2 Segmentation Step (Fig. 1B)
Registration and Subset Restriction. Each template MRI is nonlinearly registered to the test MRI to increase shape similarity (Fig. 1B-1). We used ANIMAL non-linear registration tool [13], enhanced with a boundary-based similarity measure [14]. Registration was based on a volume-of-interest that includes the labels of all hippocampi in the template library, plus a margin of 10 voxels in each direction to account for additional shape variability. Applying the registration to the library surface \( \widehat{{S^{t} }} \), it is placed on the test MRI. We then re-compute patch features across vertices and compare these patch features \( \widehat{{P_{v}^{t} }} \) with the template library patch distribution, using vertex-wise z-scoring:
This is an element-wise operation. The absolute deviation from the library can be quantified by summing the squared norm of each patch over all vertices through:
where K is the number of vertices. Figure 1B-2 shows vertex-wise deviation maps. Surfaces in the template library are ranked according to this measure, with smaller scores indicating better fit. To obtain an initial estimation, we a performed successive surface averaging (Fig. 1B-3) defined as:
where l corresponds to the ranking index. To sum up, \( \overline{{S^{1} }} \) corresponds the best template, \( \overline{{S^{2} }} \) to the weighted surface average of the two best templates and \( \overline{{S^{k} }} \) to the weighted surface average of the k best templates. Corresponding deviation scores \( \overline{{F^{k} }} \) were computed as in (2) and (3), and the one resulting in the minimal measure is chosen as initialization for a deformable model. This selection of templates has the advantage to automatically adapt to the template library size.
Deformable Model.
To further increase segmentation accuracy and to account for potential errors in the preceding steps, we applied parametric deformable model of the surface average \( \overline{S} \) [15]. The use of an explicit parameterization of the surface ensures vertex-wise correspondence across the library that would be otherwise lost (e.g. when using level-sets [16]). The objective function to minimize is composed of a regularization term, based on mechanical properties of the surface (stretching and bending), and a data term, which is represented by the deviation score F. Surface deformation is performed using gradient descent search:
where \( \overrightarrow {{x_{v} }} \) represents the spatial coordinates of voxel v, γ is the step size controlling the magnitude of the surface’s deformation; α and β are parameters controlling for surface stretching and bending. S (2) v and S (4) v are surface’s second and fourth order spatial derivative respectively at voxel v. ΔF v represents the gradient of the surface’s deviation score at vertex v. Figure 2B-4 illustrates a final segmentation.
3 Experiments and Results
3.1 Material
The training set includes 25 subjects from a public repositoryFootnote 1 (31 ± 7 yrs, 13 females) of MRI and manually-drawn labels (CA1-3, DG-CA4, SUB; average intra-/inter-rater Dice >90/87 %.) [11]. MRI data consist of isotropic T1-weighted millimetric (1 mm3) and submillimetric (0.6 mm3) 3D-MPRAGE and anisotropic 2D T2-weighted TSE (0.4 × 0.4 × 2 mm3). Images underwent automated correction for intensity non-uniformity [17] and intensity standardization. Submillimetric data was resampled to 0.4 mm3 resolution in MNI152 space. The patient cohort consists of 17 TLE patients. MRI post-processing followed the same steps as in [11]. TLE diagnosis and lateralization of the seizure focus was based on a multi-disciplinary evaluation. Hippocampal atrophy was determined as hippocampal volumes beyond 2SD of the corresponding mean of healthy controls [18].
3.2 Experiments
Parameter optimization and robustness to library size were performed on submillimetric T1-weighted images.
Parameter Optimization.
Parameters for the active contour are empirically set to α = 100 and β = 100. The step size parameter γ is set to 10−5. Performance with regards to patch sizes was evaluated using a leave-one-out (LOO) strategy, based on Dice overlap index between automated/manual segmentations.
Robustness to Template Library Variations and Image Resolution.
For each subject, we randomly decreased the library from the full size (n = 24 in LOO validation) to 1/2 (12), 1/3 (8) and 1/5 (5) of its original size. We repeated this process 5 times. We evaluated performance with smaller template libraries, based on Dice overlaps. We tested whether SurfPatch achieved adequate performance by operating solely on standard 1 mm3 MPRAGE data. In this evaluation, we first linearly upsampled images to 0.4 mm3, followed by the segmentation outlined above. This permitted the use of equivalent patch sizes. In addition to Dice, we computed correlation coefficients between automated as well as manual volumes, and generated Bland-Altman plots.
TLE Lateralization.
Direct dice overlap comparisons between SurfPatch and both ASHS and FreeSurfer are challenged by the absence of a unified subfield segmentation protocol and by the optimization of different algorithms to different MRI sequences. We thus assessed the clinical utility of the different approaches using a “TLE lateralization challenge” that assessed the accuracy of linear discriminant analysis (LDA) classifiers to lateralize the seizure focus in individual patients based on subfields volumes obtained with SurfPatch compared to those using volumes generated by FreeSurfer 5.3Footnote 2 [7] and ASHSFootnote 3 [10]. We ran both algorithms with their required modalities (FreeSurfer: 1 mm3 T1-weighted; ASHS: 0.4 × 0.4 × 2 mm3 T2-weighted) and default parameters. As both algorithms operate in native space, subfields volumes were corrected for intracranial volume by multiplying them by the Jacobian determinant of the corresponding linear transform to MNI152 space. Cross-validation was performed using a 5-Fold scheme, repeated 200 times.
ASHS Evaluation.
Given that it includes an atlas building tool, we also trained ASHS using our template library. Inputs are submillimetric T1-weighted and T2-weighted images, resampled to MNI152 space along with the corresponding labels. T1-weighted images are used for registration and T2-weighted images for segmentation.
3.3 Results
Parameter Optimization and Robustness to Template Library Size.
Maximum accuracy was achieved with a patch size of 13 × 13 × 13 voxels for CA1-3 (% Dice: 87.43 ± 2.47), 19 × 19 × 19 for CA4-DG (82.71 ± 2.85) and 11 × 11 × 11 for SUB (84.95 ± 2.45) (Fig. 2A). Mean Dice indices remained >80 % for all structures when using only 8 templates (Fig. 2B).
Robustness with Respect to Standard T1-Weighted Images.
Segmenting subfields using only standard millimetric T1-weighted images, we obtained accuracy of 85.71 ± 2.48 for CA1-3 (average decrease compared to submillimetric T1-MRI = −1.72 %), 81.10 ± 3.86 for DG (−1.61 %) and 82.21 ± 3.72 for SUB (−2.75 %). We obtained overall higher correlations between manual and automated volumes for submillimetric (Fig. 3A) than for standard images (Fig. 3B; submillimetric/millimetric CA1-3: 0.73/0.64, CA4-DG: 0.44/0.28, SUB: 0.56/0.63). Bland-Altman plots suggested lower bias in submillimetric than standard images (average shrinkage based on submillimetric/millimetric images for CA1-3: 58/131 mm3 (1.6/3.6 % from average manual volume), CA4-DG: 23/83 mm3 (3.4/8.3 %), SUB: 76/35 mm3 (4.2/1.9 %)). Segmentation examples with SurfPatch are shown in Fig. 4.
TLE Lateralization.
Lateralization of the seizure focus in TLE patients was highly accurate when using SurfPatch, both based on submillimetric and millimetric T1-weighted images (>93 %; Table 1). For ASHS and FreeSurfer, we performed two experiments using: (i) single subfields as defined by the anatomical templates and (ii) subfields grouped into CA1-3, DG-CA4 and SUB, as in [11]. Although better results were obtained with the second option, overall performance was lower than with SurfPatch (Table 1).
ASHS Evaluation.
Trained on our library, ASHS achieved similar performance as SurfPatch (CA1-3: 87.36 ± 1.97; CA4-DG: 82.54 ± 3.45; SUB: 85.48 ± 2.43).
4 Discussion and Conclusion
SurfPatch is a novel subfield segmentation algorithm combining surface-based processing with patch similarity measures. Its use of a population-based patch normalization relative to a template library has desirable run-time and space complexity properties. Moreover, it operates on T1-weighted images only, the currently preferred anatomical contrast of many big data MRI initiatives, and thus avoids T2-weighted MRI, a modality prone to motion and flow artifacts.
In controls, accuracy was excellent, with Dice overlap indices of >82 % when submillimetric images were used and only marginal performance drops when using millimetric data. Performance remained robust when reducing the size of the template library, an advantageous feature given high demands on expertise/time for the generation of subfield-specific atlases. While Dice indices across studies need to be cautiously interpreted given differences in protocols, our results compare favorably to the literature. Indeed, FreeSurfer achieved 62 % for CA1, 74 % in CA2-3 and 68 % in DG-CA4 when applied to high-resolution T1-MRI [7]. With respect to ASHS, slightly lower Dice indices than for our evaluations have been previously reported [10], particularly for CA (80 %) and SUB (75 %), whereas similarly high performance was achieved for DG (82 %). It is possible that the reliance of ASHS on anisotropic images presents a challenge to cover shape variability in antero-posterior direction. It has to be noted that ASHS achieved similar performance than SurfPatch, when trained on our library and dataset.
Although ASHS, FreeSurfer and SurfPatch consistently achieved high lateralization performance, learners based on volume measures derived from the latter lateralized the seizure focus more accurately than the other two. Robust performance on diseased hippocampi may stem from the combination of the patch-based framework, offering intrinsic modeling of multi-scale intensity features with surface-based feature sampling, which may more flexibly capture shape deformations and displacements seen in this condition.
Notes
- 1.
Data available at: http://www.nitrc.org/projects/mni-hisub25.
- 2.
FreeSurfer freely available at: http://freesurfer.net/fswiki/DownloadAndInstall.
- 3.
ASHS and UPenn PMC atlas freely available at: https://www.nitrc.org/projects/ashs/.
References
Blumcke, I., Thom, M., Aronica, E., Armstrong, D.D., Bartolomei, F., Bernasconi, A., et al.: International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: a task force report from the ILAE commission on diagnostic methods. Epilepsia 54(7), 1315–1329 (2013)
Kim, H., Mansi, T., Bernasconi, N., Bernasconi, A.: Surface-based multi-template automated hippocampal segmentation: application to temporal lobe epilepsy. Med. Image Anal. 16(7), 1445–1455 (2012)
Giraud, R., Ta, V.T., Papadakis, N., Manjon, J.V., Collins, D.L., Coupe, P., et al.: An optimized PatchMatch for multi-scale and multi-feature label fusion. Neuroimage 124(Pt A), 770–782 (2016)
Buades, A., Coll, B., Morel, J.M.: A review of image denoising algorithms, with a new one. Multiscale Model Sim. 4(2), 490–530 (2005)
Manjon, J.V., Coupe, P., Buades, A., Fonov, V., Louis Collins, D., Robles, M.: Non-local MRI upsampling. Med. Image Anal. 14(6), 784–792 (2010)
Winterburn, J.L., Pruessner, J.C., Chavez, S., Schira, M.M., Lobaugh, N.J., Voineskos, A.N., et al.: A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. Neuroimage 74, 254–265 (2013)
Van Leemput, K., Bakkour, A., Benner, T., Wiggins, G., Wald, L.L., Augustinack, J., et al.: Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus 19(6), 549–557 (2009)
Iglesias, J.E., Augustinack, J.C., Nguyen, K., Player, C.M., Player, A., Wright, M., et al.: A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: application to adaptive segmentation of in vivo MRI. Neuroimage 115, 117–137 (2015)
Pipitone, J., Park, M.T., Winterburn, J., Lett, T.A., Lerch, J.P., Pruessner, J.C., et al.: Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage 101, 494–512 (2014)
Yushkevich, P.A., Pluta, J.B., Wang, H., Xie, L., Ding, S.L., Gertje, E.C., et al.: Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum. Brain Mapp. 36(1), 258–287 (2015)
Kulaga-Yoskovitz, J., Bernhardt, B.C., Hong, S.-J., Mansi, T., Liang, K.E., van der Kouwe, A.J.W., et al.: Multi-contrast submillimetric 3 Tesla hippocampal subfield segmentation protocol and dataset. Sci. Data 2, 150059 (2015)
Styner, M., Oguz, I., Xu, S., Brechbuhler, C., Pantazis, D., Levitt, J.J., et al.: Framework for the statistical shape analysis of brain structures using SPHARM-PDM. Insight J. 1071, 242–250 (2006)
Collins, D.L., Holmes, C.J., Peters, T.M., Evans, A.C.: Automatic 3-D model-based neuroanatomical segmentation. Hum. Brain Mapp. 3(3), 190–208 (1995)
Greve, D.N., Fischl, B.: Accurate and robust brain image alignment using boundary-based registration. Neuroimage 48(1), 63–72 (2009)
Kass, M., Witkin, A., Terzopoulos, D.: Snakes – active contour models. Int. J. Comput. Vision 1(4), 321–331 (1987)
Osher, S., Sethian, J.A.: Fronts propagating with curvature-dependent speed - algorithms based on Hamilton-Jacobi formulations. J. Comput. Phys. 79(1), 12–49 (1988)
Sled, J.G., Zijdenbos, A.P., Evans, A.C.: A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging 17(1), 87–97 (1998)
Bernasconi, N., Bernasconi, A., Caramanos, Z., Antel, S.B., Andermann, F., Arnold, D.L.: Mesial temporal damage in temporal lobe epilepsy: a volumetric MRI study of the hippocampus, amygdala and parahippocampal region. Brain. 126(2), 462–469 (2003)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Caldairou, B., Bernhardt, B.C., Kulaga-Yoskovitz, J., Kim, H., Bernasconi, N., Bernasconi, A. (2016). A Surface Patch-Based Segmentation Method for Hippocampal Subfields. In: Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9901. Springer, Cham. https://doi.org/10.1007/978-3-319-46723-8_44
Download citation
DOI: https://doi.org/10.1007/978-3-319-46723-8_44
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46722-1
Online ISBN: 978-3-319-46723-8
eBook Packages: Computer ScienceComputer Science (R0)