Abstract
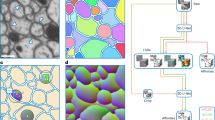
Automatic counting of neurons in fluorescently stained microscopic images is increasingly important for brain research when big imagery data sets are becoming a norm and will be more so in the future. In this paper, we present an automatic learning-based method for effective detection and counting of neurons with stained nuclei. A shape map that reflects the boosted edge and shape information is generated and a learning problem is formulated to detect the centers of stained nuclei. The method combines multiple cues of edge gradient, shape, and texture during shape map generation, feature extraction and final count determination. The proposed algorithm consistently delivers robust count ratios and precision rates on neurons in mouse and rat brain images that are shown to be better than alternative unsupervised and supervised counting methods.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Lin, G., Adiga, U., Olson, K., Guzowski, J.F., Barnes, C.A., Roysam, B.: A hybrid 3D watershed algorithm incorporating gradient cues and object models for automatic segmentation of nuclei in confocal image stacks. Cytom. A 56, 23–36 (2003)
Oberlaendera, M., Dercksenb, V.J., Eggera, R., Genselb, M., Sakmanna, B., Hegeb, H.-C.: Automated three-dimensional detection and counting of neuron somata. J. Neurosci. Methods 180, 147–160 (2009)
Zhou, J., Peng, H.: Counting cells in 3D confocal images based on discriminative models. In: ACM Conference on Bioinformatics, Computational Biology and Biomedicine (ACM BCB) (2011)
Sanders, J., Singh, A., Sterne, G., Ye, B., Zhou, J.: Learning-guided automatic three dimensional synapse quantification for drosophila neurons. BMC Bioinform. 16, 1–13 (2015)
Sobel, I., Feldman, G.: A 3×3 isotropic gradient operator for image processing. In: The Stanford Artificial Intelligence Project (SAIL) (1968)
Mallat, S.: A Wavelet Tour of Signal Processing. Academic, San Diego (1999)
Chang, C.-C., Lin, C.-J.: LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 1–27 (2011)
Peng, H., Ruan, Z., Long, F., Simpson, J.H., Myers, E.W.: V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 28, 348–353 (2010)
Acknowledgements
We thank Dr. Dragan Maric for providing the image for Bioimage Informatics Conference 2015 Nucleus Counting Challenge. The work was partially supported by NIH NIMH R15 MH099569 (Zhou) and R21 NS094091 from NIH and a Seed Grant from the Brain Research Foundation (Ye).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Ekstrom, A., Suvanto, R.W., Yang, T., Ye, B., Zhou, J. (2016). Robust Neuron Counting Based on Fusion of Shape Map and Multi-cue Learning. In: Ascoli, G., Hawrylycz, M., Ali, H., Khazanchi, D., Shi, Y. (eds) Brain Informatics and Health. BIH 2016. Lecture Notes in Computer Science(), vol 9919. Springer, Cham. https://doi.org/10.1007/978-3-319-47103-7_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-47103-7_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-47102-0
Online ISBN: 978-3-319-47103-7
eBook Packages: Computer ScienceComputer Science (R0)