Abstract
Earth observation from satellite sensory data pose challenging problems, where machine learning is currently a key player. In recent years, Gaussian Process (GP) regression and other kernel methods have excelled in biophysical parameter estimation tasks from space. GP regression is based on solid Bayesian statistics, and generally yield efficient and accurate parameter estimates. However, GPs are typically used for inverse modeling based on concurrent observations and in situ measurements only. Very often a forward model encoding the well-understood physical relations is available though. In this work, we review three GP models that respect and learn the physics of the underlying processes in the context of inverse modeling. First, we will introduce a Joint GP (JGP) model that combines in situ measurements and simulated data in a single GP model. Second, we present a latent force model (LFM) for GP modeling that encodes ordinary differential equations to blend data-driven modeling and physical models of the system. The LFM performs multi-output regression, adapts to the signal characteristics, is able to cope with missing data in the time series, and provides explicit latent functions that allow system analysis and evaluation. Finally, we present an Automatic Gaussian Process Emulator (AGAPE) that approximates the forward physical model via interpolation, reducing the number of necessary nodes. Empirical evidence of the performance of these models will be presented through illustrative examples of vegetation monitoring and atmospheric modeling.
G. Camps-Valls—The research was funded by the European Research Council (ERC) under the ERC-CoG-2014 SEDAL project (grant agreement 647423), and the Spanish Ministry of Economy and Competitiveness (MINECO) through the project TIN2015-64210-R.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Earth observation
- Remote sensing
- Vegetation
- Kernel methods
- Gaussian processes
- Inverse modeling
- Geosciences
- Transfer models
1 Introduction
Solving inverse problems is a recurrent topic of research in Physics in general, and in Earth Observation (EO) in particular. Earth Observation encompasses geosciences, climate science and remote sensing. After all, Science is about making inferences about physical parameters from sensory data. A very relevant inverse problem is that of estimating vegetation properties from remotely sensed images. Accurate inverse models help to determine the phenological stage and health status (e.g., development, productivity, stress) of crops and forests [12], which has important societal, environmental and economical implications. Leaf chlorophyll content (Chl), leaf area index (LAI), and fractional vegetation cover (FVC) are among the most important vegetation parameters to retrieve from space observations [15, 24].
In general, mechanistic models implement the laws of Physics and allow us to compute the data values given a model [21]. This is known as the forward problem. In the inverse problem, the aim is to reconstruct the model from a set of measurements, see Fig. 1. Notationally, a forward model describing the system is expressed as \(\mathbf {x}=g(\mathbf {y},\varvec{\omega })\), where \(\mathbf {x}\) is a measurement obtained by the satellite (e.g. radiance); the vector \(\mathbf {y}\) represents the state of the biophysical variables on the Earth (which we desire to infer or predict and is often referred to as outputs in the inverse modeling approach); \(\varvec{\omega }\) contains a set of controllable conditions (e.g. wavelengths, viewing direction, time, Sun position, and polarization); and \(g(\cdot )\) is a function which relates \(\mathbf {y}\) with \(\mathbf {x}\). Such a function g is typically considered to be nonlinear, smooth and continuous. Our goal is to obtain an inverse model, \(f(\cdot ) \approx g^{-1}(\cdot )\), parametrized by \(\varvec{\theta }\), which approximates the biophysical variables \(\mathbf {y}\) given the data \(\mathbf {x}\) received by the satellite, i.e.  . Radiative transfer models (RTMs) are typically used to implement the forward direction [13, 22]. However, inverting RTMs directly is very complex because the number of unknowns is generally larger than the number of independent radiometric information [14]. Also, estimating physical parameters from RTMs is hampered by the presence of high levels of uncertainty and noise, primarily associated to atmospheric conditions and sensor calibration. This translates into inverse problems where deemed similar spectra may correspond to very diverse solutions. This gives raise to undetermination and ill-posed problems.
. Radiative transfer models (RTMs) are typically used to implement the forward direction [13, 22]. However, inverting RTMs directly is very complex because the number of unknowns is generally larger than the number of independent radiometric information [14]. Also, estimating physical parameters from RTMs is hampered by the presence of high levels of uncertainty and noise, primarily associated to atmospheric conditions and sensor calibration. This translates into inverse problems where deemed similar spectra may correspond to very diverse solutions. This gives raise to undetermination and ill-posed problems.
Methods for model inversion and parameter retrieval can be roughly separated in three main families: statistical, physical and hybrid methods [10]. Statistical inversion predicts a biogeophysical parameter of interest using a training dataset of input-output data pairs coming from concurrent measurements of the parameter of interest (e.g. leaf area index -LAI-) and the corresponding satellite observations (e.g. reflectances). Statistical methods typically outperform other approaches, but ground truth measurements involving a terrestrial campaign are necessary. Physical inversion reverses RTMs by searching for similar spectra in look-up-tables (LUTs) and assigning the parameter corresponding to the most similar observed spectrum. This requires selecting an appropriate cost function, and generating a rich, representative LUT from the RTM. The use of RTMs to generate data sets is a common practice, and especially convenient because acquisition campaigns are very costly in terms of time, money, and human resources, and usually limited in terms of parameter combinations. Finally, hybrid inversion exploits the input-output data generated by RTM simulations and train statistical regression models to invert the RTM model. Hybrid models combine the flexibility and scalability of machine learning while respecting the physics encoded in the RTMs. Currently, kernel machines in general [8], and Bayesian non-parametric approaches such as Gaussian Process (GP) regression [19] in particular, are among the preferred regression models [9, 23].
While hybrid inversion is practical when no in situ data is available, intuitively it makes sense to let predictions be guided by actual measurements whenever they are present. Likewise, when only very few real in situ measurements are available, it is sensible to incorporate simulated data from RTMs to properly ground the models. This is the first pathway considered in this paper, which extends the hybrid inversion by proposing a statistical method that performs nonlinear and nonparametric inversion blending both real and simulated data. The so-called joint GP (JGP) essentially learns how to trade off noise variance in the real and simulated data.
A second topic covered in this paper follows an alternative pathway to learn latent functions that generated the observations using GP models. We introduce a latent force model (LFM) for GP modelling [1]. The proposed LFM-GP combines the ordinary differential equations of the forward model (through smoothing kernels) and empirical data (from in situ campaigns). The LFM presented here performs multi-output structured regression, adapts to the signal characteristics, is able to cope with missing data in the time series, and provides explicit latent functions that allow system analysis and evaluation.
Finally, we deal with the important issue of emulation, that is learning surrogate GP models to approximate costly RTMs. The proposed Automatic Gaussian Process Emulator (AGAPE) methodology combines the interpolation capabilities of Gaussian processes (GPs) with the accurate design of an acquisition function that favours sampling in low density regions and flatness of the interpolation function.
2 Gaussian Process Models for Inverse Modeling
GPs are state-of-the-art tools for regression and function approximation, and have been recently shown to excel in biophysical variable retrieval by following both statistical [9, 23] and hybrid approaches [6, 7]. Let us consider a set of n pairs of observations or measurements, \({\mathcal D}_n:=\{\mathbf {x}_i, y_i\}_{i=1}^n\), perturbed by an additive independent noise. The input data pairs \((\mathbf {X},\mathbf {y})\) used to fit the inverse machine learning model \(f(\cdot )\) come from either in situ field campaign data (statistical approach) or simulations by means of an RTM (hybrid approach). We assume the following model,
where \(f(\mathbf {x})\) is an unknown latent function, \(\mathbf {x}\) \(\in \) \({\mathbb R}^d\), and \(\sigma _n^2\) stands for the noise variance. Defining \(\mathbf {y}\) \(=\) \([y_1, \ldots , y_n]^\intercal \) and \(\mathbf {f}\) \(=\) \([f(\mathbf {x}_1),\ldots , f(\mathbf {x}_n)]^\intercal \), the conditional distribution of \(\mathbf {y}\) given \(\mathbf {f}\) becomes \(p(\mathbf {y}| \mathbf {f}) = \mathcal {N}(\mathbf {f}, \sigma _n^2\mathbf {I})\), where \(\mathbf {I}\) is the \(n\times n\) identity matrix. Now, in the GP approach, we assume that \(\mathbf {f}\) follows a n-dimensional Gaussian distribution \(\mathbf {f} \sim \mathcal {N}(\varvec{0}, \mathbf {K})\) [3].
The covariance matrix \(\mathbf{K}\) of this distribution is determined by a kernel function with entries \(\mathbf {K}_{ij}=k(\mathbf {x}_i,\mathbf {x}_j)=\exp (-\Vert \mathbf {x}_i-\mathbf {x}_j\Vert ^2/(2\sigma ^2))\), encoding similarity between the input points [19]. The intuition here is the following: the more similar input i and j are, according to some metric, the more correlated output i and j ought to be. Thus, the marginal distribution of \(\mathbf {y}\) can be written as
where \(\varvec{C}_n=\mathbf {K}+ \sigma _n^2\mathbf {I}\). Now, what we are really interested in is predicting a new output \(y_*\), given an input \(x_*\) (Fig. 2). The GP framework handles this by constructing a joint distribution over the training and test points,
where \(\mathbf {k}_{*} = [k(\mathbf {x}_*,\mathbf {x}_1), \ldots , k(\mathbf {x}_*,\mathbf {x}_n)]^\intercal \) is an \(n\times 1\) vector and \(c_*= k(\mathbf {x}_*,\mathbf {x}_*) + \sigma _n^2\). Then, using standard GP manipulations, we can find the distribution over \(y_*\) conditioned on the training data, which is a normal distribution with predictive mean and variance given by
Thus, GPs yield not only predictions \(\mu _{\text {GP}*}\) for test data, but also the so-called “error-bars”, \(\sigma _{\text {GP}*}\), assessing the uncertainty of the mean prediction. The hyperparameters \(\varvec{\theta }=[\sigma , \sigma _n]\) to be tuned in the GP determine the width of the squared exponential kernel function and the noise on the observations. This can be done by marginal likelihood maximization or simple grid search, attempting to minimize the squared prediction errors.
3 Forward and Inverse Joint GP Models
Let us assume that the previous dataset \({\mathcal D}_n\) is formed by two disjoint sets: one set of r real data pairs, \({\mathcal D}_r=\{(\mathbf {x}_i, y_i)\}_{i=1}^r\), and one set of s RTM-simulated pairs \({\mathcal D}_s=\{(y_j,\mathbf {x}_j)\}_{j=r+1}^n\), so that \(n=r+s\) and \({\mathcal D}_n={\mathcal D}_r\cup {\mathcal D}_s\). In matrix form, we have \(\mathbf {X}_r\in {\mathbb R}^{r\times d}\), \(\mathbf {y}_r\in {\mathbb R}^{r\times 1}\), \(\mathbf {X}_s\in {\mathbb R}^{s\times d}\) and \(\mathbf {y}_s\in {\mathbb R}^{s\times 1}\), containing all the inputs and outputs of \({\mathcal D}_r\) and \({\mathcal D}_s\), respectively. Finally, the \(n\times 1\) vector \(\mathbf {y}\) contains all the n outputs, sorted with the real data first, followed by the simulated data. Now, we define a different model, where the observation noise depends on the origin of the data: \(\sigma _n^2\) for real observations (\(\mathbf {x}_i \in {\mathcal D}_r\)) or \(\sigma _n^2/\gamma \) for RTM simulations (\(\mathbf {x}_i \in {\mathcal D}_s\)), where the parameter \(\gamma >0\) accounts for the importance of the two sources of information relative to each other.
The resulting distribution of \(\mathbf {y}\) given \(\mathbf {f}\) is only slightly different from that of the regular GP, namely \(p(\mathbf {y}|\mathbf {f}) = \mathcal {N}(\mathbf {f}, \sigma _n^2 \mathbf {V})\) where \(\mathbf {V}\) is an \(n\times n\) diagonal matrix in which the first r diagonal elements are equal to 1 and the remaining s are equal to \(\gamma ^{-1}\): \(\mathbf {V}=\text {diag}(1,\ldots ,1,\gamma ^{-1},\ldots , \gamma ^{-1})\). The predictive mean and variance of a test output \(y_*\), conditioned on the training data, then becomes
Note that when \(\gamma =1\) the standard GP formulation is obtained. Otherwise \(\gamma \) acts as an extra regularization term accounting for the relative importance of the real and the simulated data points. The hyperparameters of the JGP are \(\varvec{\theta }= [\sigma ,\sigma _n,\gamma ],\) which can be selected by maximizing the marginal likelihood of the observations as usual in the GP framework [19]. It is important to note that hyperparameter fitting should be performed with respect to real data, so that the method learns the mapping from real input to output.
3.1 Experimental Results
We are concerned with the prediction of leaf area index (LAI) parameter from space, a parameter that characterizes plant canopies and is roughly defined as the total needle surface area per unit ground area. Non-destructive real LAI data were acquired over Elementary Sampling Units (ESUs) within rice fields in Spain, Italy and Greece during field campaigns in 2015 and 2016. The temporal frequency of the campaigns was approximately 10 days starting from the very beginning of rice emergence (early-June) up to the maximum rice green LAI development (mid-August). LAI measurements were acquired using a dedicated smartphone app (PocketLAI), which uses both the smartphone’s accelerometer and camera to acquire images at 57.5\(^\circ \) below the canopy and computes LAI through an internal segmentation algorithm [7]. The center of the ESU was geo-located for later matching and association of the mean LAI estimate with the corresponding satellite spectra. We used Landsat 8 surface reflectance data over each area corresponding to the dates of measurements’ acquisition. The resulting datasets contain a number of in situ measurements in the range of 70-300 depending on the country and year. On the other hand, a simulated data set of \(s=2000\) pairs of Landsat 8 spectra and LAI was obtained running the PROSAIL RTM in forward mode.Footnote 1 The leaf and canopy variables as well as the soil brightness parameter, were generated following a PROSAIL site-specific parameterization to constrain the model to Mediterranean rice areas [6].
We assessed the performance of JGP for different amounts of real and simulated data. The gain in accuracy was measured as the reduction in root mean square error (RMSE gain [%] = \(100\,\times \,\)(RMSE\(_{\text {GP}}\)-RMSE\(_{\text {JGP}}\))/RMSE\(_{\text {GP}}\)). We evaluated performance in the 6 datasets generated for different countries (SP, GR, IT) and years (2015, 2016). Figure 3 shows the effect of the ratio between simulated and real data points \(p = s/r\) on the RMSE gain evaluated using 10-fold crossvalidation. When no simulated data is used, the JGP model reduces to the standard GP model, but when introducing an amount of PROSAIL-datapoints similar to the amount of real datapoints, i.e. \(p\sim 1\), a noticeable gain is for datasets gathered in 2016. In the case of the data from Spain, the gain appears rather stable (between 6 and 2% in 2015 and 2016 respectively) after reaching a ratio of \(p=2\), indicating what size of the simulated dataset is needed for an increase in accuracy. The results for Greece and Italy, however, show that the use of simulated data attempting to fill in the under-represented domain of the real data, is not always useful.
4 Inverse Modelling with Latent Force Models
We are interested in inverse modelling from real in situ data, and to learn not only an accurate retrieval model but also about the physical mechanism that generated the input-output observed relations without even accessing any RTM, see Fig. 4. Here, we assume that our observations correspond simply to the temporal variable, \(\mathbf {x}\sim t\), so the latent functions are defined in the time domain, \(f_r(t)\). Nevertheless, extension to multidimensional objects such as radiances is straighforward by using kernels. Notationally, let us consider a multioutput scenario with Q correlated observed time series, \(y_q(t)\) for \(1 \le q \le Q\), and let us assume that we have n samples available for each of these signals, taken at sampling points \(t_i\), s.t. \(y_q[i] = y_q(t_i)\) for \(1 \le i \le n\). This is the training set, which is composed of an input vector, \(\mathbf {t}=[t_1,\ \ldots ,\ t_n]^{\top }\), and an output matrix, \(\mathbf {Y}=[\mathbf {y}_1,\ \ldots ,\ \mathbf {y}_Q]\) with \(\mathbf {y}_q=[y_q[1],\ \ldots ,\ y_q[n]]^{\top }\). We aim to build a GP model for the Q outputs that can be used to perform inference on the test set: \(\widetilde{\mathbf {t}}=[\widetilde{t}_1,\ \ldots ,\ \widetilde{t}_m]^{\top }\) and \(\widetilde{\mathbf {Y}}=[\widetilde{\mathbf {y}}_1,\ \ldots ,\ \widetilde{\mathbf {y}}_Q]\) with \(\widetilde{\mathbf {y}}_q=[\widetilde{y}_q[1],\ \ldots ,\ \widetilde{y}_q[m]]^{\top }\) and \(\widetilde{y}_q[m'] = y_q(\widetilde{t}_{m'})\) for test inputs at \(t_{m'}\).
Formulation. Let us assume that a set of R independent latent functions (LFs), \(f_r(t)\) with \(1 \le r \le R\), are responsible for the observed correlation between the outputs. Then, the cross-correlation between the outputs arises naturally as a result of the coupling between the set of independent LFs, instead of being imposed directly on the set of outputs. Let us define the form of these latent functions and the coupling mechanism between them. In this work, we model the LFs as zero-mean Gaussian processes (GPs), and the coupling system emerges through a linear convolution operator described by an impulse response, \(h_{q}(t)\), as follows:
where \(\text {L}_{q}[t]\left\{ f_r(t)\right\} \) indicates the linear operator associated to the linear convolution. The resulting outputs are finally obtained as a linear weighted combination of these pseudo-outputs plus an additive white Gaussian noise (AWGN) term:
where \(S_{r,q}\) represents the coupling strength between the r-th LF and the q-th output, and \(w_q(t) \sim \mathcal {N}(0,\eta _q^2)\) is the AWGN term. In practice, we consider only the squared exponential auto-covariance function for the LFs, \(k_{f_r f_r}(t'-t) \propto \exp (-\frac{(t'-t)^2}{2 \ell _r^2})\), where the hyperparameter \(\ell _r\) controls the length-scale of the process. The smoothing kernel encodes our knowledge about the linear system (that relates the unobserved LFs and the outputs), and can be based on basic physical principles of the system at hand (as in [1]) or selected arbitrarily (as in [4, 11]). In this paper, we consider also the Gaussian smoothing kernel, \(h_q(t) \propto \exp (-\frac{t^2}{2 \nu _q^2})\). Now, since the LFs are zero-mean GPs, the noise is also zero-mean and Gaussian, and all the operators involved are linear, the joint LFs-outputs process is also a GP. Therefore, the mean function of the q-th output is \(\mu _{y_q}(t) = 0\), whereas the cross-covariance function between two outputs is
where the term \(\text {L}_{p}[t]\left\{ \text {L}_{q}[t']\left\{ k_{f_r f_r}(t,t')\right\} \right\} \) denotes the application of the convolutional operator twice to the autocorrelation function of the LFs, which results in the following double integral:
Finally, the cross-correlation between the LFs and the outputs readily gives \(k_{f_r y_q}(t,t') = S_{r,q} \text {L}_{q}[t']\left\{ k_{f_r f_r}(t,t')\right\} \), which involves a single one-dimensional integral already computed in an intermediate step before. All integrals can be solved analytically when both the LFs and the smoothing kernel have a Gaussian shape.
Learning hyperparameters is very challenging through marginal log-likelihood maximization because of its complicated dependence on hyperparameters \(\varvec{\theta }=[\nu _q,l_r,\sigma ,\sigma _n,\eta _q]\). We propose to solve the problem through a stochastic gradient descent technique, the scaled conjugate gradient [18]. Once the hyperparameters \(\varvec{\theta }\) of the model have been learned, inference proceeds by applying standard GP regression formulas [19] (cf. Sect. 2). Now, since the conditional PDF is Gaussian, the minimum mean squared error (MMSE) prediction is simply given by the conditional mean:
where \(\hat{\mathbf {y}} = [\hat{\mathbf {y}}_1^{\top },\ \ldots ,\ \hat{\mathbf {y}}_Q^{\top }]\) is the vectorized version of the inferred outputs, which can be expressed in matrix form as \(\hat{\mathbf {Y}}=[\hat{\mathbf {y}}_1,\ \ldots ,\ \hat{\mathbf {y}}_Q]\) with \(\hat{\mathbf {y}}_q=[\hat{y}_q[1],\ \ldots ,\ \hat{y}_q[m]]^{\top }\) and \(\hat{y}_q[m'] = \hat{y}_q(\widetilde{t}_m')\).
4.1 Experimental Results
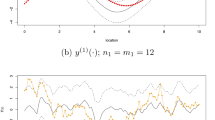
We are concerned about multiple time series of two (related) biophysical parameters, LAI and fAPAR (Fraction of Absorbed Photosynthetically Active Radiation), in the locations of the experiments in Sect. 3.1. We focus on a set of representative rice pixels of each area, thus allowing us to observe the inter-annual variability of rice from 2003 to 2014 at a coarse spatial resolution (2 Km), which is useful for regional vegetation modelling. We focus on learning the latent forces for the multi-output time series composed of the LAI and fAPAR data for Spain and Italy (i.e., the number of outputs is \(Q=4\)) from the beginning of 2003 until the end of 2013. We use all the LAI data available from the MODIS sensor for Spain (\(N=506\) samples), and the first half (years 2003–2009) of the other three time series. The recovered LF (\(R=1\)) and two examples of the modelled time series are displayed in Fig. 5. Note that the model has succeeded in capturing the dynamics of the data by using a single LF. Good numerical results were obtained: for Spain, we have MSE = 0.1139 and MSE = 0.0080 for LAI and fAPAR respectively, whereas for Italy we have MSE = 0.2422 and MSE = 0.0046, respectively.
Gap filling example using a single LF (i.e., \(R=1\)). Training used all the LAI data from Spain (years 2003–2013) and the first half (years 2003–2009) of the other three time series: fAPAR (ES), LAI (IT) and fAPAR (IT). The second half constitutes the test set of such time series. Training data (red circles), test data (red dashed line), predicted time series (black line) and uncertainty measured by \(\pm 2\) standard deviations about the mean predicted value (gray shaded area). (Color figure online)
5 Automatic GP Emulation
Emulation deals with the challenging problem of building statistical models for complex physical RTMs. The emulators are also called surrogate or proxy models, and they try to learn the equations encoded from data. Namely, an emulator is a statistical model which tries to reproduce the behavior of a deterministic and very costly physical model. Emulators built with GPs are gaining popularity in remote sensing and geosciences, since they allow efficient data processing and sensitivity analysis [5, 9, 20]. Here, we are interested in optimizing emulators such that a minimal number of simulations is run. The technique is called AGAPE (automatic Gaussian Process emulator), and is related to some Bayesian optimization and active learning techniques.
The goal is to interpolate a costly function \(g(\mathbf {y})\) choosing adequately the nodes, in order to reduce the error in the interpolation with the smallest possible number of evaluation of \(g(\mathbf {y})\). Given an input matrix of nodes (used for the interpolation) at the t-th iterations, \(\mathbf {Y}_t=[\mathbf {y}_1\cdots \mathbf {y}_{m_t}]\), of dimension \(d\times m_t\) (where d is the dimension of each \(\mathbf {y}_i\) and \(m_t\) is the number of points), we have a vector of outputs, \(\mathbf{x}_t=[x_1,\ldots ,x_{m_t}]^{\intercal }\), where \(x_t=g(\mathbf {y}_t)\) is the estimation of the observations (e.g., radiances) at iteration \(t\in \mathbb {N}^+\) of the algorithm. Figure 6 shows a graphical representation of a generic automatic emulator. At each iteration t one performs an interpolation, \(\widehat{g}_t(\mathbf {y}|\mathbf {Y}_t,\mathbf {x}_t)\), followed by an optimization step that updates the acquisition function, \(A_{t}(\mathbf {y})\), updates the set \(\mathbf {Y}_{t+1} =[\mathbf {Y}_t, \mathbf {y}_{m_t+1}]\) adding a new node, set \(m_t \leftarrow m_t +1\) and \(t\leftarrow t+1\). The procedure is repeated until a suitable stopping condition is met, such as a certain maximum number of points is included or a desired precision error \(\epsilon \) is achieved, \(\Vert \widehat{g}_t(\mathbf {y})-\widehat{g}_{t-1}(\mathbf {y})\Vert \le \epsilon \).
Formulation. the acquisition function, \(A_t(\mathbf {y})\), encodes useful information for proposing new points to build the emulator. At each iteration, a new node is added maximizing \(A_t(\mathbf {y})\), i.e.,
and set \(\mathbf {Y}_{t+1}=[\mathbf {Y}_t,\mathbf {y}_{m_{t}+1}]\), \(m_{t+1}=m_t+1\). Here, we propose to account for both a geometry \(G_t(\mathbf {y})\) and a diversity \(D_t(\mathbf {y})\),
where \(A_{t}(\mathbf {y}): \mathcal {Y}\mapsto \mathbb {R}\), and \(\beta _t\) is an increasing function with respect to t, with \(\textstyle \lim _{t\rightarrow \infty } \beta _t=1\) (or \(\beta _t=1\) for \(t>t'\)). Function \(G_t(\mathbf {y})\) captures the geometrical information in g, while function \(D_t(\mathbf {x})\) depends on the distribution of the points in the current vector \(\mathbf {Y}_t\). More specifically, \(D_t(\mathbf {y})\) will have a greater probability mass around empty areas within \(\mathcal {Y}\), whereas \(D_t(\mathbf {y})\) will be approximately zero close to the support points and exactly zero at the support points, i.e., \(D_t(\mathbf {y}_i)=0\), for \(i=1,\ldots ,m_t\) and \(\forall t\in \mathbb {N}\). Since g is unknown, the function \(G_t(\mathbf {y})\) can be only derived from information acquired in advance or by considering the approximation \(\widehat{g}\). The tempering value, \(\beta _t\), helps to downweight the likely less informative estimates in the very first iterations. If \(\beta _t=0\), we disregard \(G_t(\mathbf {y})\) and \(A_{t}(\mathbf {y})= D_t(\mathbf {y})\), whereas, if \(\beta _t=1\), we have \(A_{t}(\mathbf {y})=G_t(\mathbf {y}) D_t(\mathbf {y})\).
We consider a GP for emulation, so the inputs and outputs are now reversed. In addition, note that interpolation fixes \(\sigma _n=0\). Therefore, the AGAPE predictive mean and variance at iteration t for a new point \(\mathbf {y}_*\) become simply
where now \(\mathbf {k}_{*} = [k(\mathbf {y}_*,\mathbf {y}_1), \ldots , k(\mathbf {y}_*,\mathbf {y}_{m_t})]^\intercal \) contains the similarities between the input point \(\mathbf {x}_*\) and the observed ones at iteration t, \(\mathbf {K}\) is an \(m_t\times m_t\) kernel matrix with entries \(\mathbf {K}_{i,j}:=k(\mathbf {y}_i,\mathbf {y}_j)\), and \(\varvec{\alpha }=\mathbf {K}_{nn}^{-1}\mathbf {x}_t\) is the coefficient vector for interpolation. The interpolation for \(\mathbf {y}_*\) can be simply expressed as a linear combination of \(\widehat{g}_t(\mathbf {y}_*) = \mathbf{k}_*^\intercal {\varvec{\alpha }} = \sum _{i=1}^{m_t} \alpha _i k(\mathbf {y}_*,\mathbf {y}_i)\).
Note that \(\sigma _{\text {AGAPE}}^2(\mathbf {y}_i)=0\) for all \(i=1,\ldots , m_t\) and \(\sigma _{\text {AGAPE}}^2(\mathbf {y})\) depends on the distance among the support points \(\mathbf {y}_t\), and the chosen kernel function k and associated hyper-parameter \(\sigma \). For this reason, the function \(\sigma _{\text {AGAPE}}^2(\mathbf {y})\) is a good candidate to represent the distribution of the \(\mathbf {y}_t\)’s since it is zero at each \(\mathbf {y}_i\), and higher far from the points \(\mathbf {y}_i\)’s. Moreover, \(\sigma _{\text {AGAPE}}^2(\mathbf {y})\) takes into account the information of the GP interpolator. Therefore, we consider as the diversity term \(D(\mathbf {y}):=\sigma _{\text {AGAPE}}^2(\mathbf {y})\), i.e., \(D(\mathbf {y})\) is induced by the GP interpolator.
As geometric information, we consider enforcing flatness on the interpolation function, and thus aim to minimize the norm of the the gradient of the interpolating function \(\widehat{g}_t\) w.r.t. the input data \(\mathbf {y}\), i.e., \(G(\mathbf {y}) = \Vert \nabla _y\widehat{g}_t(\mathbf {y}|\mathbf {Y}_t,\mathbf {x}_t)\Vert = \Vert \sum _{i=1}^{m_t} \alpha _i\nabla _y k(\mathbf {y},\mathbf {y}_i)\Vert \). This intuitively makes wavy regions of g require more support points than flat regions. The gradient vector for the squared exponential kernelm \(k(\mathbf {y},\mathbf {y}')=\exp (-\Vert \mathbf {y}-\mathbf {y}'\Vert ^2/(2\sigma ^2))\) with \(\mathbf {y}=[y_1,\ldots ,y_d]^{\intercal }\), can be computed in closed-form, \(\nabla _y k(\mathbf {y},\mathbf {y}')=-\frac{k(\mathbf {y},\mathbf {y}')}{\sigma ^2}[(y_1-y'_1),\ldots ,(y_d-y'_d)]^{\intercal },\) so the geometry term \(G(\mathbf {y})\) can be defined, for instance, as follows:
which reduces the dependence to the current approximation \(\widehat{g}_t\). Therefore, the acquisition function can be readily obtained by defining \(\beta _t=1-\exp (-\gamma t)\), where \(\gamma \ge 0\) is a positive scalar and plugging Eq. (9) into Eq. (8). We optimized \(A(\mathbf {x})\) using interacting parallel simulated annealing methods [16, 17].
5.1 Experimental Results
We show empirical evidence of performance on the optimization of selected points for a complex and computationally expensive RTM: the MODTRAN5-based LUT. MODTRAN5 is considered as the de facto standard atmospheric RTM for atmospheric correction applications [2]. In our test application, and for the sake of simplicity, we have considered \(d=2\) with the Aerosol Optical Thickness at 550 nm (\(\tau \)) and ground elevation (h) as key input parameters. The underlying function \(g(\mathbf {y})\) consists therefore on the execution of MODTRAN5 at given values of \(\tau \) and h and wavelength of 760 nm. The input parameter space is bounded to 0.05–0.4 for \(\tau \) and 0–3 km for h. In order to test the accuracy of the different schemes, we have evaluated \(g(\mathbf {y})\) at all the possible 1750 combinations of 35 values of \(\tau \) and 50 values of h. Namely, this thin grid represents the ground-truth in this example.
We tested (a) a standard, yet suboptimal, random approach choosing points uniformly within \(\mathcal {Y}=[0.05,0.4]\times [0,3]\), (b) the Latin Hypercube sampling [5], and (c) the proposed AGAPE. We start with \(m_0=5\) points \(\mathbf {y}_1=[0.05,0]^{\top }\), \(\mathbf {y}_2=[0.05,3]^{\top }\), \(\mathbf {y}_3=[0.4,0]^{\top }\), \(\mathbf {y}_4=[0.4,3]^{\top }\) and \(\mathbf {y}_5=[0.2,1.5]^{\top }\) for all the techniques. We compute the final number of nodes \(m_t\) required to obtain an \(\ell _2\) distance between g and \(\widehat{g}\) smaller than \(\epsilon =0.03\), with the different methods. The results, averaged over \(10^3\) runs, are shown in Table 1. AGAPE requires the addition of \(\approx 4\) new points to obtain a distance smaller than 0.03.
6 Conclusions
We introduced three different schemes based on GP modeling in the interplay between Physics and Machine Learning, with the focus on the Earth system modeling. Canonical machine learning for EO problems rely on in situ observational data, and often disregard the physical knowledge and models available. We argue that the equations encoded in forward physical models may be very useful in inverse GP modeling, such that models may give consistent, physically meaningful estimates. Three types of physics-aware GP models were introduced: a simple approach to combine in situ measurements and simulated data in a single GP model, a latent force model that incorporates ordinary differential equations, and an automatic compact emulator of physical models through GPs. The developed models demonstrated good performance, adaptation to the signal characteristics and transportability to unseen situations.
Notes
- 1.
PROSAIL simulates leaf reflectance for the optical spectrum, from 400 to 2500 nm with a 1 nm spectral resolution, as a function of biochemistry and structure of the canopy, its leaves, the background soil reflectance and the sun-view geometry.
References
Álvarez, M.A., Luengo, D., Lawrence, N.D.: Linear latent force models using gaussian processes. IEEE Trans. Patt. Anal. Mach. Intell. 35(11), 2693–2705 (2013)
Berk, A., Anderson, G., Acharya, P., Bernstein, L., Muratov, L., Lee, J., Fox, M., Adler-Golden, S., Chetwynd, J., Hoke, M., Lockwood, R., Gardner, J., Cooley, T., Borel, C., Lewis, P., Shettle, E.: MODTRAN5: 2006. In: International Society for Optics and Photonics (2006)
Bishop, C.M.: Pattern recognition. Mach. Learn. 128, 1–58 (2006)
Boyle, P., Frean, M.: Dependent gaussian processes. In: NIPS, pp. 217–224 (2004)
Busby, D.: Hierarchical adaptive experimental design for gaussian process emulators. Reliab. Eng. Syst. Saf. 94, 1183–1193 (2009)
Campos-Taberner, M., García-Haro, F., Camps-Valls, G., Grau-Muedra, G., Nutini, F., Crema, A., Boschetti, M.: Multitemporal and multiresolution leaf area index retrieval for operational local rice crop monitoring. Rem. Sens. Environ. 187, 102–118 (2016)
Campos-Taberner, M., Garcia-Haro, F., Moreno, A., Gilabert, M., Sanchez-Ruiz, S., Martinez, B., Camps-Valls, G.: Mapping leaf area index with a smartphone and gaussian processes. IEEE Geosci. Remote Sens. Lett. 12(12), 2501–2505 (2015)
Camps-Valls, G., Bruzzone, L. (eds.): Kernel Methods for Remote Sensing Data Analysis. Wiley, UK (2009)
Camps-Valls, G., Muñoz-Marí, J., Verrelst, J., Mateo, F., Gomez-Dans, J.: A survey on gaussian processes for earth observation data analysis. IEEE Geosci. Remote Sens. Mag. 3(2), 1–20 (2016)
Camps-Valls, G., Tuia, D., Gómez-Chova, L., Jiménez, S., Malo, J. (eds.): Remote Sens. Image Process. Morgan & Claypool Publishers, USA (2011)
Higdon, D.: Space and space-time modeling using process convolutions. In: Anderson, C.W., Barnett, V., Chatwin, P.C., El-Shaarawi, A.H. (eds.) Quantitative Methods for Current Environmental Issues, pp. 37–54. Springer, London (2002)
Hilker, T., Coops, N.C., Wulder, M.A., Black, T.A., Guy, R.D.: The use of remote sensing in light use efficiency based models of gross primary production: a review of current status and future requirements. Sci. Total. Environ. 404(2–3), 411–423 (2008)
Jacquemoud, S., Bacour, C., Poilvé, H., Frangi, J.P.: Comparison of four radiative transfer models to simulate plant canopies reflectance: direct and inverse mode. Remote Sens. Environ. 74(3), 471–481 (2000)
Liang, S.: Advances in Land Remote Sensing: System, Modeling, Inversion and Applications. Springer, Germany (2008)
Lichtenthaler, H.K.: Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382 (1987)
Martino, L., Elvira, V., Luengo, D., Corander, J., Louzada, F.: Orthogonal parallel MCMC methods for sampling and optimization. Dig. Sign Proc. 58, 64–84 (2016)
Read, J., Martino, L., Luengo, D.: Efficient monte carlo optimization for multi-label classifier chain. In: IEEE International Conference on Acoustics, Speech, and Signal Processing, ICASSP, pp. 1–5 (2013)
Nabney, I.: NETLAB: Algorithms for Pattern Recognition. Springer, London (2002)
Rasmussen, C.E., Williams, C.K.I.: Gaussian Processes for Machine Learning. The MIT Press, New York (2006)
Rivera, J., Verrelst, J., Gómez-Dans, J., Muñoz-Marí, J., Moreno, J., Camps-Valls, G.: An emulator toolbox to approximate radiative transfer models with statistical learning. Remote Sens. 7(7), 9347–9370 (2015)
Snieder, R., Trampert, J.: Inverse Problems in Geophysics. Springer, Vienna (1999)
Verhoef, W., Bach, H.: Simulation of hyperspectral and directional radiance images using coupled biophysical and atmospheric radiative transfer models. Remote Sens. Environ. 87, 23–41 (2003)
Verrelst, J., Alonso, L., Camps-Valls, G., Delegido, J., Moreno, J.: Retrieval of vegetation biophysical parameters using gaussian process techniques. IEEE Trans. Geosci. Remote Sens. 50(5), 1832–1843 (2012)
Whittaker, R.H., Marks, P.L.: Methods of assessing terrestrial productivity. In: Lieth, H., Whittaker, R.H. (eds.) Primary Productivity of the Biosphere, vol. 14, pp. 55–118. Springer, Heidelberg (1975)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Camps-Valls, G. et al. (2017). Physics-Aware Gaussian Processes for Earth Observation. In: Sharma, P., Bianchi, F. (eds) Image Analysis. SCIA 2017. Lecture Notes in Computer Science(), vol 10270. Springer, Cham. https://doi.org/10.1007/978-3-319-59129-2_18
Download citation
DOI: https://doi.org/10.1007/978-3-319-59129-2_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-59128-5
Online ISBN: 978-3-319-59129-2
eBook Packages: Computer ScienceComputer Science (R0)