Abstract
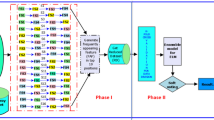
Feature/Gene selection is a major research area in the study of gene expression data, generally dealing with classification tasks of diseases or subtype of diseases and identification of biomarkers related to a type of disease. In such a context, this paper proposes an ensemble approach of gene selection for classification tasks from gene expression datasets. This proposal provides a four-staged approach of gene filtering. Each stage performs a different gene filtering task, such as: data processing, noise removing, gene selection ensemble and application of wrapper methods to reach the end result, a small subset of informative genes. Our proposal has been assessed on two different datasets of the same disease (Pancreatic ductal adenocarcinoma) for which, good results have been achieved in comparison with other gene selection methods. Hence, the proposed strategy has proven its reliability with respect to other approaches.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Badea, L., Herlea, V., Olimpia, S., Dumitrascu, T., Popescu, I.: Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology 88, 2015–2026 (2008)
Kota, J., Hancock, J., Kwon, J., Korc, M.: Pancreatic cancer: stroma and its current and emerging targeted therapies. Cancer Lett. 391, 38–49 (2017)
Bhaw-Luximon, A., Jhurry, D.: New avenues for improving pancreatic ductal adenocarcinoma (pdac) treatment: selective stroma depletion combined with nano drug delivery. Cancer Lett. 369(2), 266–273 (2015)
Korc, M.: Pancreatic cancer-associated stroma production. Am. J. Surg. 194(4), S84–S86 (2007). Elsevier
Hidalgo, M., Cascinu, S., Kleeff, J., Labianca, R., Löhr, J.M., Neoptolemos, J., Real, F.X., Van Laethem, J.L., Heinemann, V.: Addressing the challenges of pancreatic cancer: future directions for improving outcomes. Pancreatology 15(1), 8–18 (2015). Elsevier
Natarajan, A., Ravi, T.: A survey on gene feature selection using microarray data for cancer classification. Int. J. Comput. Sci. Commun. (IJCSC) 5(1), 126–129 (2014)
Shraddha, S., Anuradha, N., Swapnil, S.: Feature selection techniques and microarray data: a survey. Int. J. Emerg. Technol. Adv. Eng. 4(1), 179–183 (2014)
Tyagi, V., Mishra, A.: A survey on different feature selection methods for microarray data analysis. Int. J. Comput. Appl. 67(16), 36–40 (2013)
Castellanos-Garzón, J.A., Ramos, J.: A gene selection approach based on clustering for classification tasks in colon cancer. Adv. Distrib. Comput. Artif. Intell. J. (ADCAIJ) 4(3), 1–10 (2015). http://dx.doi.org/10.14201/ADCAIJ201543110
Hezel, A., Kimmelman, A., Stanger, B., Bardeesy, N., DePinho, R.: Genetics and biology of pancreatic ductal adenocarcinoma. Genes & Dev. 20, 1218–1249 (2006)
Fang, Z., Du, R., Cui, X.: Uniform approximation is more appropriate for wilcoxon rank-sum test in gene set analysis. PLoS ONE 7(2), e31505 (2012)
Weiss, P.: Applications of generating functions in nonparametric tests. Math. J. 9(4), 803–823 (2005)
Lazar, C., Taminau, J., Meganck, S., Steenhoff, D., Coletta, A., Molter, C., deSchaetzen, V., Duque, R., Bersini, H., Nowé, A.: A survey on filter techniques for feature selection in gene expression microarray analysis. IEEE/ACM Trans. Comput. Biol. Bioinform. 9(4) 1106–1118 (2012)
Berrar, D.P., Dubitzky, W., Granzow, M.: A Practical Approach to Microarray Data Analysis. Kluwer Academic Publishers, New York (2003)
Wolters, M.: A genetic algorithm for selection of fixed-size subsets with application to design problems. J. Stat. Softw. 68(1), 1–18 (2015)
Kursa, M., Rudnicki, W.: Feature selection with the Boruta package. J. Stat. Softw. 36(11), 1–13 (2010)
Mahmoud, O., Harrison, A., Perperoglou, A., Gul, A., Khan, Z., Metodiev, M., Lausen, B.: A feature selection method for classification within functional genomics experiments based on the proportional overlapping score. BMC Bioinform. 15(274), 1–20 (2014)
Ahdesmaki, A., Strimmer, K.: Feature selection in omics prediction problems using CAT scores and false non-discovery rate control. Ann. Appl. Stat. 4, 503–519 (2010)
Ishwaran, H., Rao, J.: Spike and slab variable selection: frequentist and bayesian strategies. Ann. Stat. 33(2), 730–773 (2005)
Friedman, J., Hastie, T., Tibshirani, R.: Regularization paths for generalized linear modelsvia coordinate descent. J. Stat. Softw. 33(1), 1–22 (2008). http://www.stanford.edu/~hastie/Papers/glmnet.pdf
Acknowledgments
This work has been supported by project MOVIURBAN: Máquina social para la gestión sostenible de ciudades inteligentes: movilidad urbana, datos abiertos, sensores móviles. SA070U 16. Project co-financed with Junta Castilla y León, Consejería de Educación and FEDER funds.
The research of Daniel López-Sánchez has been financed by the Ministry of Education, Culture and Sports of the Spanish Government (University Faculty Training (FPU) program, reference number FPU15/02339).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Castellanos-Garzón, J.A., Ramos, J., López-Sánchez, D., de Paz, J.F. (2017). An Ensemble Approach for Gene Selection in Gene Expression Data. In: Fdez-Riverola, F., Mohamad, M., Rocha, M., De Paz, J., Pinto, T. (eds) 11th International Conference on Practical Applications of Computational Biology & Bioinformatics. PACBB 2017. Advances in Intelligent Systems and Computing, vol 616. Springer, Cham. https://doi.org/10.1007/978-3-319-60816-7_29
Download citation
DOI: https://doi.org/10.1007/978-3-319-60816-7_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-60815-0
Online ISBN: 978-3-319-60816-7
eBook Packages: EngineeringEngineering (R0)