Abstract
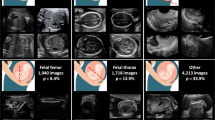
Accurate classification and localization of anatomical structures in images is a precursor for fully automatic image-based diagnosis of placental abnormalities. For placental ultrasound images, typically acquired in clinical screening and risk assessment clinics, these structures can have quite indistinct boundaries and low contrast, and image-level interpretation is a challenging and time-consuming task even for experienced clinicians. In this paper, we propose an automatic classification model for anatomy recognition in placental ultrasound images. We employ deep residual networks to effectively learn discriminative features in an end-to-end fashion. Experimental results on a large placental ultrasound image database (10,808 distinct 2D image patches from 60 placental ultrasound volumes) demonstrate that the proposed network architecture design achieves a very high recognition accuracy (0.086 top-1 error rate) and provides good localization for complex anatomical structures around the placenta in a weakly supervised fashion. To our knowledge this is the first successful demonstration of multi-structure detection in placental ultrasound images.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bazzani, L., Bergamo, A., Anguelov, D., Torresani, L.: Self-taught object localization with deep networks. In: 2016 IEEE Winter Conference on Applications of Computer Vision (WACV), pp. 1–9. IEEE (2016)
Chen, H., Dou, Q., Yu, L., Heng, P.A.: Voxresnet: deep voxelwise residual networks for volumetric brain segmentation. arXiv preprint arXiv:1608.05895 (2016)
Collins, S.L., Ashcroft, A., Braun, T., Calda, P., Langhoff-Roos, J., Morel, O., Stefanovic, V., Tutschek, B., Chantraine, F.: Proposal for standardized ultrasound descriptors of abnormally invasive placenta (AIP). Ultrasound Obstet. Gynecol. 47(3), 271–275 (2016)
Collins, S.L., Stevenson, G.N., Al-Khan, A., Illsley, N.P., Impey, L., Pappas, L., Zamudio, S.: Three-dimensional power doppler ultrasonography for diagnosing abnormally invasive placenta and quantifying the risk. Obstet. Gynecol. 126(3), 645–653 (2015)
Collins, S., Stevenson, G., Noble, J., Impey, L., Welsh, A.: Influence of power doppler gain setting on virtual organ computer-aided analysis indices in vivo: can use of the individual sub-noise gain level optimize information? Ultrasound Obstet. Gynecol. 40(1), 75–80 (2012)
Collobert, R., Kavukcuoglu, K., Farabet, C.: Torch7: A matlab-like environment for machine learning. In: BigLearn, NIPS Workshop (2011)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
He, K., Zhang, X., Ren, S., Sun, J.: Identity mappings in deep residual networks. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9908, pp. 630–645. Springer, Cham (2016). doi:10.1007/978-3-319-46493-0_38
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167 (2015)
Kamnitsas, K., Ledig, C., Newcombe, V.F., Simpson, J.P., Kane, A.D., Menon, D.K., Rueckert, D., Glocker, B.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Oquab, M., Bottou, L., Laptev, I., Sivic, J.: Is object localization for free?-weakly-supervised learning with convolutional neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 685–694 (2015)
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., Huang, Z., Karpathy, A., Khosla, A., Bernstein, M., Berg, A.C., Fei-Fei, L.: ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. (IJCV) 115(3), 211–252 (2015)
Simonyan, K., Vedaldi, A., Zisserman, A.: Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034 (2013)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Srivastava, N., Hinton, G.E., Krizhevsky, A., Sutskever, I., Salakhutdinov, R.: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15(1), 1929–1958 (2014)
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning deep features for discriminative localization. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 2016
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Qi, H., Collins, S., Noble, A. (2017). Weakly Supervised Learning of Placental Ultrasound Images with Residual Networks. In: Valdés Hernández, M., González-Castro, V. (eds) Medical Image Understanding and Analysis. MIUA 2017. Communications in Computer and Information Science, vol 723. Springer, Cham. https://doi.org/10.1007/978-3-319-60964-5_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-60964-5_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-60963-8
Online ISBN: 978-3-319-60964-5
eBook Packages: Computer ScienceComputer Science (R0)