Abstract
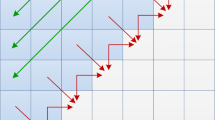
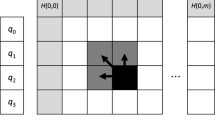
The well-known Smith-Waterman (SW) algorithm is the most commonly used method for local sequence alignments. However, SW is very computationally demanding for large protein databases. There are several implementations that take advantage of parallel capacities on many-cores, FPGAs or GPUs, in order to increase the alignment throughtput. In this paper, we have explored SW acceleration on Intel KNL processor. The novelty of this architecture requires the revision of previous programming and optimization techniques on many-core architectures. To the best of authors knowledge, this is the first KNL architecture assessment for SW algorithm. Our evaluation, using the renowned Environmental NR database as benchmark, has shown that multi-threading and SIMD exploitation showed competitive performance (351 GCUPS) in comparison with other implementations.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Environmental NR: ftp://ftp.ncbi.nih.gov/blast/db/FASTA/env_nr.gz.
- 2.
Swiss-Prot: http://web.expasy.org/docs/swiss-prot_guideline.html.
- 3.
SSE4.1 and AVX2 versions using QP technique were excluded from the analysis to improve figure readability since we found that SP scheme always achieved the best performance, as in previous work [13].
References
Asai, R.: MCDRAM as high-bandidth memory (HBM) in knights landing processors: developer’s guide (2016). https://goparallel.sourceforge.net/wp-content/uploads/2016/05/Colfax_KNL_MCDRAM_Guide.pdf
Daily, J.: Parasail: SIMD C library for global, semi-global, and local pairwise sequence alignments. BMC Bioinform. 17, 81 (2016)
Gotoh, O.: An improved algorithm for matching biological sequences. J. Mol. Biol. 162, 705–708 (1981)
Isa, M., Benkrid, K., Clayton, T., Ling, C., Erdogan, A.: An FPGA-based parameterised and scalable optimal solutions for pairwise biological sequence analysis. In: 2011 NASA/ESA Conference on Adaptive Hardware and Systems (AHS), pp. 344–351, June 2011
Lan, H., Liu, W., Schmidt, B., Wang, B.: Accelerating large-scale biological database search on xeon phi-based neo-heterogeneous architectures. In: 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 503–510, November 2015
Li, T.I., Shum, W., Truong, K.: 160-fold acceleration of the Smith-Waterman algorithm using a field programmable gate array (FPGA). BMC Bioinform. 8, I85 (2007)
Liu, Y., Wirawan, A., Schmidt, B.: CUDASW++ 3.0: accelerating Smith-Waterman protein database search by coupling CPU and GPU SIMD instructions. BMC Bioinform. 14, 117 (2013)
Liu, Y., Schmidt, B.: Swaphi: Smith-waterman protein database search on xeon phi coprocessors. In: 25th IEEE International Conference on Application-Specific Systems, Architectures and Processors (ASAP 2014) (2014)
Mount, D.W.: Bioinformatics: Sequence and Genome Analysis. Mount Bioinformatics. Cold Spring Harbor Laboratory Press, New York (2004)
Reinders, J., Jeffers, J., Sodani, A.: Intel Xeon Phi Processor High Performance Programming Knights, Landing edn. Morgan Kaufmann Publishers Inc., Boston (2016)
Rognes, T.: Faster Smith-Waterman database searches with inter-sequence SIMD parallelisation. BMC Bioinform. 12(1), 221 (2011). http://dx.doi.org/10.1186/1471-2105-12-221
Rognes, T., Seeberg, E.: Six-fold speed-up of Smith-Waterman sequence database searches using parallel processing on common microprocessors. Bioinformatics 16(8), 699 (2000). http://dx.doi.org/10.1093/bioinformatics/16.8.699
Rucci, E., Garcia, C., Botella, G., De Giusti, A., Naiouf, M., Prieto-Matas, M.: An energy-aware performance analysis of SWIMM: Smith Waterman implementation on Intel’s Multicore and Manycore architectures. Concurr. Comput. Pract. Exp. 27(18), 5517–5537 (2015). http://dx.doi.org/10.1002/cpe.3598
Rucci, E., Garcia, C., Botella, G., De Giusti, A., Naiouf, M., Prieto-Matas, M.: OSWALD: OpenCL Smith-Waterman algorithm on altera FPGA for large protein databases. Int. J. High Perform. Comput. Appl. (2016). http://dx.doi.org/10.1177/1094342016654215
Smith, T.F., Waterman, M.S.: Identification of common molecular subsequences. J. Mol. Biol. 147(1), 195–197 (1981)
Sodani, A., Gramunt, R., Corbal, J., Kim, H.S., Vinod, K., Chinthamani, S., Hutsell, S., Agarwal, R., Liu, Y.C.: Knights landing: second-generation intel xeon phi product. IEEE Micro 36(2), 34–46 (2016)
Acknowledgments
This work has been partially supported by Spanish government through research contract TIN2015-65277-R and CAPAP-H6 network (TIN2016-81840-REDT).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Rucci, E., Garcia, C., Botella, G., De Giusti, A., Naiouf, M., Prieto-Matias, M. (2017). First Experiences Accelerating Smith-Waterman on Intel’s Knights Landing Processor. In: Ibrahim, S., Choo, KK., Yan, Z., Pedrycz, W. (eds) Algorithms and Architectures for Parallel Processing. ICA3PP 2017. Lecture Notes in Computer Science(), vol 10393. Springer, Cham. https://doi.org/10.1007/978-3-319-65482-9_42
Download citation
DOI: https://doi.org/10.1007/978-3-319-65482-9_42
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-65481-2
Online ISBN: 978-3-319-65482-9
eBook Packages: Computer ScienceComputer Science (R0)