Abstract
Delineation of the lung and lobar anatomy in MR images is challenging due to the limited image contrast and the absence of visible interlobar fissures. Here we propose a novel automated lung and lobe segmentation method for pulmonary MR images. This segmentation method employs prior information of the lungs and lobes extracted from CT in the form of multiple MRI atlases, and adopts a learning-based atlas-encoding scheme, based on random forests, to improve the performance of multi-atlas segmentation. In particular, we encode each CT-derived MRI atlas by training an atlas-specific random forest for each structure of interest. In addition to appearance features, we also extract label context features from the registered atlases to introduce additional information to the non-linear mapping process. We evaluated our proposed framework on 10 clinical MR images acquired from COPD patients. It outperformed state-of-the-art approaches in segmenting the lungs and lobes, yielding a mean Dice score of 95.7%.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Accurate segmentation of the lungs and their lobes is an essential prerequisite for quantitative regional analysis of lung structure and function. Manual segmentation performed by an expert is still regarded as the gold standard for most clinical applications, despite its limited reproducibility and labour-intensive nature. Although many studies for automatic lung and lobe segmentation have been reported in the thoracic computed tomography (CT) literature [2, 9], there has been very little analogous work in MR imaging (MRI). This is mainly hampered by the limited definition of relevant structures arising from intrinsic low proton density in pulmonary airspaces, and the absence of fine structures such as fissures that separate the lungs into lobes.
As pioneering work on automatic MRI-based lung segmentation, Lelieveldt et al. [7] introduced a model-matching approach to extract the lungs by registering the image to an anatomical model built from a manually annotated dataset. In a more recent attempt, Osareh and Shadgar [8] applied a region-aided geodesic active contour model in conjunction with fuzzy c-means clustering to delineate the lung cavities. Kohlmann et al. [6] tackled the problem by introducing an automatic multi-step lung segmentation pipeline based on histogram thresholding, region growing, and morphological operations. Recently, Guo et al. proposed semi-automatic convex optimisation-based methods that incorporated the inherent left-to-right lung volume proportion prior [4] and image features from proton and hyperpolarised helium-3 MRI [5]. Even though these segmentation methods have shown promising results in conventional lung MRI, there are still limitations that need to be addressed prior to clinical implementation including efficient workflow and improved segmentation performance. For instance, some of these approaches required user interaction [4, 5], while others were heavily reliant on robust grey-level thresholding [6], or restricted to 2-D MR image applications [8]. In addition, none of these proposed methodological frameworks are able to segment the lung lobes for more localised regional analysis. This hinders the establishment of an MRI-only regional lung function evaluation protocol without the need for a CT scan.
To tackle the aforementioned limitations, we introduce a novel learning-based multi-atlas segmentation (MAS) framework based on random forests to segment the lung and lobar anatomy in proton MRI. Specifically, we leverage the prior information from a database of CT images in the form of CT-based MRI atlases to compensate for the comparatively reduced structural information in MRI. We then encode each atlas by training a set of atlas-specific random forests (one classifier per label or structure of interest (SOI)) using both appearance and atlas label context features to delineate each lung structure. In this way we aim at overcoming the limitations of previous MAS and learning-based approaches. The main contributions of this work are: (1) the development of an automatic pipeline for segmenting the lungs and lobes in proton MRI; (2) the introduction of a novel atlas encoding scheme based on random forests into a MAS framework; (3) a validation of our proposed method with comparison to state-of-the-art approaches.
2 Materials and Methods
2.1 Image Data
The dataset used comes from 10 patients diagnosed with chronic obstructive pulmonary disease (COPD), each with corresponding CT and proton MRI scans at a single time point. The scans were part of a study involving hyperpolarised gas MRI acquisition, and this introduced some requirements on the protocol; however, only the standard proton MR and CT images were used for the purpose of this paper. MR images were obtained on a 1.5 T Signa HDx whole-body MR system using a balanced steady-state free precession sequence (resolution = 1.56\(\,\times \,\)1.56\(\,\times \,\)15 mm\(^{3}\)), while CT images were acquired after 60 seconds post-intravenous contrast on a 16-slice GE Discovery 670 scanner (resolution = 0.7\(\,\times \,\)0.7\(\,\times \,\)1.25 mm\(^{3}\)). Both MRI and CT acquisition were carried out during a 15-second breath-hold after inhalation of 1 L oxygen via a Tedlar bag from functional residual capacity. Manual segmentation of the lobes in CT and lungs in MRI were performed using the ITK-SNAP software (www.itksnap.org). The former was exploited to generate a CT-based MRI atlas library, whereas the latter was used as the gold standard for validation of lung segmentation in MRI.
2.2 Construction of a CT-Based Lung MRI Atlas Library
Each CT image was first spatially registered to its corresponding MR image using rigid and affine transforms followed by a diffeomorphic B-spline transform, with mutual information as similarity metric. The manually annotated CT label maps were then warped to the corresponding MR image using the resulting transformations. This process was repeated for each of the 10 CT images resulting in 10 lung MRI atlases with prior information of the lungs and lobes extracted from CT. These atlases formed the atlas library used in this work.
2.3 Learning-Based Multi-atlas Segmentation
To enforce spatial consistency and flexibility, we propose to train an atlas-specific random forest for each atlas’s label or SOI. This setting also helps to combat unbalanced classes in the training data, and aggregation of predictions from different classifiers trained for different SOIs (ensemble of classifiers) could produce a better result than a single global classifier. We use random forests since they are computationally efficient and can effectively handle a great number of high-dimensional data.
Atlas Encoding: Figure 1 presents an overview of the atlas encoding scheme to learn atlas-specific random forests. Denote an atlas library as A with N atlases {\(A_{i}=(I_{i},L_{i})|i=1,...,N\)}, where \( I_{i} \) and \( L_{i} \) are respectively the intensity image and corresponding label map of the i-th atlas. To train the i-th atlas-specific forest, we first spatially aligned the i-th atlas image \(I_{i}\) to each of the other atlases, which are now treated as training targets {\(T_{k}=(D_{k},G_{k})|k=1,...,N-1\)}, where \( D_{k}\) and \(G_{k}\) are the intensity image and the label map of the k-th training target image. The resulting transformation is then applied to the i-th atlas label map \( L_{i} \). As a result, (\( N-1 \)) training pairs {\( D_{k}, I_{i}^{k}, L_{i}^{k}, G_{k} \)} are formed, and \( G_{k} \) is used as the class label for training random forests while \( D_{k} \), \(I_{i}^{k}\) and \( L_{i}^{k}\) are utilised for feature extraction. For each training pair, balanced positive and negative samples are drawn from each SOI for training a random forest. Local multi-scale appearance features are then extracted for each training sample x in the (training) target image \( D_{k}\). To extract atlas label context features, we apply 3-D PatchMatch [3] to search the top similarity patch \( n_{1}(x) \) of x within a search volume V from the registered atlas image \(I_{i}^{k}\). This step is essential to alleviate potential registration errors. We then extract label context features from the corresponding local patch of \( n_{1}(x) \) in the warped atlas label map \( L_{i}^{k}\). These two types of features are eventually concatenated and used for inferring label. After training, r x N atlas forests will be learned for r SOIs.
The flowchart of our proposed learning-based atlas encoding scheme, consisting of steps: (1) registration, (2) non-local patch selection, (3) feature extraction, and (4) classifier training. The red box indicates the image patch centered at the training voxel x, while the yellow box represents the search volume V.
Feature Extraction: We use 3-D Haar-like features [10], which have been demonstrated successfully in a number of applications including biomedical image analysis, to capture local multi-scale appearance information of lung structures. The equation for computing Haar-like features can be written as:
where P denotes the 3-D patch centred at voxel x of an image I, and \( \alpha \) is either 0 or 1, determining whether one or two cubical patches are used. Cubic patches \( P_{1} \) and \( P_{2} \) are randomly chosen from the size range {1 .. 11} and randomly displayed within a local neighbourhood P of size 11\(\,\times \,\)11\(\,\times \,\)11 voxels. On the other hand, we also extract context features from the registered atlas label map. In particular, we sparsely select 300 voxels within a neighbourhood size of 11\(\,\times \,\)11\(\,\times \,\)11 centred at voxel x to capture both short-range and long-range contextual information.
Atlas Forest Labelling: During testing, we first register all atlases to the to-be-segmented target image as in conventional MAS framework. Given a target voxel x, we use 3-D PatchMatch to find the top K atlas patches, which have similar local intensity to the target patch centred at x, in the registered atlases. Next, label features are extracted for each of these K atlas patches from the warped atlases, and combined with local appearance features of x, as input to each trained atlas-specific random forest for inferring label. K label probabilities will be obtained for the target voxel x from each atlas forest and fused using spatial-varying weighted voting [3], where higher weight is given to atlas patches with similar appearance to the target patch. The final labelling result of the target image will be the average of all labelling results from all atlas forests.
3 Experiments and Results
3.1 Experimental Setup
An image pre-processing pipeline was applied before training an atlas-specific random forest, consisting in bias field correction and histogram matching. All registration were performed using the ANTs software package with the default parameters (https://github.com/stnava/ANTs). In this experiment, 2-fold cross validation procedure was performed with 5 images each for training and testing. We trained 30 trees for each atlas-specific random forest with maximum tree depth of 20. Minimum number of samples for each leaf node was set as 5. 1000 Haar-like features were randomly sampled for training each tree. Search volume V of 15\(\,\times \,\)15\(\,\times \,\)15 and top 5 atlas patches were used.
We compared with state-of-the-art MAS methods, i.e. non-local patch-based label fusion (PBLF) [1] and joint label fusion (JLF) [12], on the same dataset. Specifically, we used 7\(\,\times \,\)7\(\,\times \,\)7 image patches to evaluate patch-wise similarity, model parameter \(\sigma \) = 2, and 9\(\,\times \,\)9\(\,\times \,\)9 search volume for the JLF algorithm, while a search volume of 15\(\,\times \,\)15\(\,\times \,\)15 and a patch size of 5\(\,\times \,\)5\(\,\times \,\)5 were applied for PBLF algorithm. Additionally, we computed the segmentation results produced by majority voting (MV) and STAPLE algorithm [13] as a baseline evaluation. To test the benefits of the combination of the multi-atlas and machine learning approaches, we compared with the segmentation performance produced by the random forest on its own. We trained a single random forest (SRF) classifier in a classical way without performing any registration with the same training parameters, and applied this trained classifier to segment target images. Haar-like features were used and spatial location of each voxel was included as an additional feature to avoid spatial inconsistency. The Dice similarity coefficient was used to quantify the match with manual segmentations.
3.2 Impact of the Number of Atlas Forests
The number of atlases used have an important effect in the segmentation performance of a conventional MAS method. We analysed the segmentation accuracy of our method for 1 to 5 atlas forests to segment the lungs and lobes. The results are presented in Table 1. Unsurprisingly, increasing the number of atlas forests increased the accuracy of the segmentation, as in conventional MAS techniques. This was probably because combining the decision of multiple atlas forests tended to correct random errors made by the individual atlas forests. However, as Table 1 shows, there was a small decrease of segmentation performance when 4 atlas forests were used. This might be due to the use of an irrelevant or dissimilar atlas that misguided the segmentation procedure, resulting in a small performance drop. In subsequent tests, we refer to the single-atlas forest as SAF and the five-atlas forest as MAF.
3.3 Comparison with Multi-atlas and Learning-Based Segmentation Methods
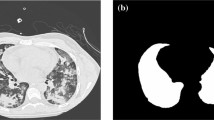
Table 2 shows the segmentation performance by all the compared methods for the lung and lobe segmentations in proton MRI. The proposed method (MAF) achieved the best performance among all the methods in all lung structures with statistical significance (\( p<0.05\)) using a paired, two-tailed t-test. It was followed by the JLF and single-atlas version of our proposed method (SAF), which outperformed the rest of the approaches (\( p<0.05\)). Although JLF was better than SAF, the differences were not statistically significant. The segmentation quality of SAF was comparable to JLF, which used all five atlases to achieve optimal performance, while only one registration was required for SAF and no computation time was needed for the combination of several candidate segmentations. For qualitative analysis, Fig. 2 shows an example of the segmentation results for the top performing algorithms.
On the other hand, the performance of the classical machine learning approach (SRF) that learned a single random forest classifier to assign labels for the entire testing image was clearly below those of multi-atlas segmentation approaches, except MV and STAPLE algorithms that ignored the image intensity information. As Fig. 2 shows, there are some misclassified voxels between lobes. By incorporating a local search volume and the patch intensities feature in label fusion, the non-local patch based label fusion (PBLF) method outperformed MV, STAPLE, and SRF, providing supportive evidence that better segmentation performance can be achieved by relaxing the one-to-one correspondence constraints.
4 Discussion and Conclusion
Combining multi-atlas and learning-based methods have been explored before in the context of random forests for brain [14] and cardiac [11] segmentation. A fundamental difference between our work and these is that we introduce a novel atlas encoding scheme that leverages both local appearance and atlas label contextual information of multiple atlases for better random forest learning. Most learning-based methods use only local intensity or appearance information, which limits the accuracy in our application, in which similar intensity patches are common. Each labelled image or atlas contains rich information about the spatial arrangement and even shape of the anatomical structures. Integrating this rich information into random forests is promising as it learns a better discriminative model, as demonstrated in our results. We also implement a non-local patch search strategy to alleviate potential atlas-target registration errors and spatial-varying weighted voting scheme in the atlas forest labelling step to improve the segmentation performance.
In conclusion, we have presented a novel learning-based multi-atlas segmentation framework based on random forests for segmenting the lungs and lobes in proton MRI with the use of prior anatomical information derived from CT. The core element of our proposed method lies in integrating the complementary advantages of both multi-atlas and machine learning based segmentation. Our method achieves state-of-the-art performance in delineating the lungs and lobes in proton MRI, though up to date this has been demonstrated only on a relatively small dataset. A comprehensive evaluation using a larger data sample will be carried out in the near future.
References
Coupe, P., Manjon, J.V., Fonov, V., Pruessner, J., Robles, M., Collins, D.L.: Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage 54(2), 940–954 (2011)
Doel, T., Gavaghan, D.J., Grau, V.: Review of automatic pulmonary lobe segmentation methods from CT. Comput. Med. Imaging Grap. 40, 13–29 (2015)
Giraud, R., Ta, V.T., Papadakis, N., Manjón, J.V., Collins, D.L., Coupé, P.: An optimized patchmatch for multi-scale and multi-feature label fusion. NeuroImage 124(Part A), 770–782 (2016)
Guo, F., Svenningsen, S., Eddy, R.L., Capaldi, D.P.I., Sheikh, K., Fenster, A., Parraga, G.: Anatomical pulmonary magnetic resonance imaging segmentation for regional structure-function measurements of asthma. Med. Phys. 43(6), 2911–2926 (2016)
Guo, F., Yuan, J., Rajchl, M., Svenningsen, S., PI Capaldi, D., Sheikh, K., Fenster, A., Parraga, G.: Globally optimal co-segmentation of three-dimensional pulmonary 1H and hyperpolarized 3He MRI with spatial consistence prior. Med. Image Anal. 23(1), 43–55 (2015)
Kohlmann, P., Strehlow, J., Jobst, B., Krass, S., Kuhnigk, J.M., Anjorin, A., Sedlaczek, O., Ley, S., Kauczor, H.U., Wielpütz, M.O.: Automatic lung segmentation method for MRI-based lung perfusion studies of patients with chronic obstructive pulmonary disease. Int. J. Comput. Assist. Radiol. Surg. 10(4), 403–417 (2015)
Lelieveldt, B.P.F., Van Der Geest, R.J., Ramze Rezaee, M., Bosch, J.G., Reiber, J.H.C.: Anatomical model matching with fuzzy implicit surfaces for segmentation of thoracic volume scans. IEEE Trans. Med. Imaging 18(3), 218–230 (1999)
Osareh, A., Shadgar, B.: A segmentation method of lung cavities using region aided geometric snakes. J. Med. Syst. 34(4), 419–433 (2010)
Rikxoort, E.M.V., Ginneken, B.V.: Automated segmentation of pulmonary structures in thoracic computed tomography scans: a review. Phys. Med. Biol. 58, R187–220 (2013)
Viola, P., Jones, M.J.: Robust real-time face detection. Int. J. Comput. Vision 57(2), 137–154 (2004)
Wang, H., Cao, Y., Syeda-Mahmood, T.: Multi-atlas segmentation with learning-based label fusion. In: Wu, G., Zhang, D., Zhou, L. (eds.) MLMI 2014. LNCS, vol. 8679, pp. 256–263. Springer, Cham (2014). doi:10.1007/978-3-319-10581-9_32
Wang, H., Suh, J.W., Das, S.R., Pluta, J.B., Craige, C., Yushkevich, P.A.: Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 35(3), 611–623 (2013)
Warfield, S.K., Zou, K.H., Wells, W.M.: Simultaneous truth and performance level estimation (staple): an algorithm for the validation of image segmentation. IEEE Trans. Med. Imaging 23(7), 903–921 (2004)
Zikic, D., Glocker, B., Criminisi, A.: Encoding atlases by randomized classification forests for efficient multi-atlas label propagation. Med. Image Anal. 18(8), 1262–1273 (2014)
Acknowledgments
HL is supported by a fellowship funded by the Ministry of Higher Education Malaysia and Universiti Malaysia Perlis. The research was partly supported by the CRUK and EPSRC Cancer Imaging Centre in Oxford.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Lee, H., Matin, T., Gleeson, F., Grau, V. (2017). Learning-Based Multi-atlas Segmentation of the Lungs and Lobes in Proton MR Images. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10433. Springer, Cham. https://doi.org/10.1007/978-3-319-66182-7_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-66182-7_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66181-0
Online ISBN: 978-3-319-66182-7
eBook Packages: Computer ScienceComputer Science (R0)