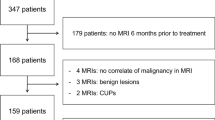
Abstract
Motion corruption can result in difficulty identifying lesions, and incorrect diagnoses by radiologists in cases of breast cancer screening using DCE-MRI. Although registration techniques can be used to correct for motion artifacts, their use has a computational cost and, in some cases can lead to a reduction in diagnostic quality rather than the desired improvement. In a clinical system it would be beneficial to identify automatically which studies have severe motion corruption and poor diagnostic quality and which studies have acceptable diagnostic quality. This information could then be used to restrict registration to only those cases where motion correction is needed, or it could be used to identify cases where motion correction fails. We have developed an automated method of estimating the degree of mis-registration present in a DCE-MRI study. We experiment using two predictive models; one based on a feature extraction method and a second one using a deep learning approach. These models are trained using estimates of deformation generated from unlabeled clinical data. We validate the predictions on a labeled dataset from radiologists denoting cases suffering from motion artifacts that affected their ability to interpret the image. By calculating a binary threshold on our predictions, we have managed to identify motion corrupted cases on our clinical dataset with an accuracy of 86% based on the area under the ROC curve. This approach is a novel attempt at defining a clinically relevant level of motion corruption.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Armato, S.G., et al.: Temporal subtraction in chest radiography: automated assessment of registration accuracy. Med. Phys. 33(5), 1239 (2006)
Klein, A., et al.: Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46(3), 786–802 (2009)
Kok, T., et al.: Efficacy of MRI and Mammography for breast-cancer screening in women with a familial or genetic predisposition. Engl. J. Med. 351(5), 427–437 (2004)
Krizhevsky, A., et al.: ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 1–9 (2012)
Muenzing, S.E.A., et al.: Supervised quality assessment of medical image registration: application to CT lung registration. Med. Image Anal. 16(8), 1521–1531 (2012)
Rueckert, D., et al.: Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18(8), 712–721 (1999)
Srinivasa Reddy, B., Chatterji, B.N.: An FFT-based technique for translation, rotation, and scale-invariant registration. IEEE Trans. Image Process. 5(8), 1266–1271 (1996)
Srivastava, N., et al.: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014)
Vignati, A., et al.: A fully automatic lesion detection method for DCE-MRI fat-suppressed breast images. SPIE Med. Imaging 7260, 726026 (2009)
Zeiler, M.D., Fergus, R.: Visualizing and understanding convolutional networks. In: Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T. (eds.) ECCV 2014. LNCS, vol. 8689, pp. 818–833. Springer, Cham (2014). doi:10.1007/978-3-319-10590-1_53
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Chiang, S., Balasingham, S., Richmond, L., Curpen, B., Skarpathiotakis, M., Martel, A. (2017). Motion Corruption Detection in Breast DCE-MRI. In: Wang, Q., Shi, Y., Suk, HI., Suzuki, K. (eds) Machine Learning in Medical Imaging. MLMI 2017. Lecture Notes in Computer Science(), vol 10541. Springer, Cham. https://doi.org/10.1007/978-3-319-67389-9_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-67389-9_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-67388-2
Online ISBN: 978-3-319-67389-9
eBook Packages: Computer ScienceComputer Science (R0)