Abstract
k-nearest neighbor (kNN) search is a fundamental data mining task critical to many data analytics methods. Yet no effective techniques to date scale kNN search to large datasets. In this work we present PkNN, an exact distributed method that by leveraging modern distributed architectures for the first time scales kNN search to billion point datasets. The key to the PkNN strategy is a multi-round kNN search that exploits pivot-based data partitioning at each stage. This includes an outlier-driven partition adjustment mechanism that effectively minimizes data duplication and achieves a balanced workload across the compute cluster. Aggressive data-driven bounds along with a tiered support assignment strategy ensure correctness while limiting computation costs. Our experimental study on multi-dimensional real-world data demonstrates that PkNN achieves significant speedup over the state-of-the-art and scales effectively in data cardinality. Code and data related to this chapter are available at: http://solar-10.wpi.edu/cakuhlman/PkNN.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Detecting the nearest neighbors of all points in a dataset is a ubiquitous task common to numerous data mining endeavors. Many techniques, including classification and regression [7], clustering [3, 10], and outlier detection [4, 20], require a k-nearest neighbor (kNN) search to be performed for every point in the dataset. As real-world applications increasingly rely on the analysis of very large datasets, they require resources beyond what is feasible on a single machine. Therefore, the development of highly distributed solutions for kNN search is no longer an option, but a necessity.
Several distributed exact kNN search methods have been proposed in the literature [5, 15, 24], using a support set strategy. First, the data is divided into disjoint partitions by grouping nearby points together. Then these core partitions are each augmented by additional sets of support points which could potentially be neighbors of the points in the core partition. The support sets ensure that a local kNN search over core points will return exact results without requiring access to data stored on any other machines.
Although this approach successfully distributes kNN computation across machines, existing methods are not scalable beyond million point datasets, as confirmed by our experiments (Sect. 5). Major shortcomings include the use of worst-case estimation for support sets [15] leading to excessive data duplication, or computationally intensive support determination leading to excessive runtimes [5]. Worse yet, the outlier problem in skewed data sets has largely been overlooked. Data points far from all others can disproportionately inflate support sets and overload machines, resulting in job failures.
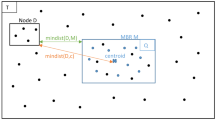
Challenges. A key property of kNN search is that the distance from each point to its neighbors may vary considerably across a dataset. The distance tends to be small in dense areas, while very large for points in sparse areas, as illustrated in Fig. 1. Therefore the distance from points to potential neighbors in the support set varies greatly across partitions. To locate the exact set of all necessary support points for each partition a priori would essentially require a kNN search be conducted even before the data partitioning is formed. Alternatively, if the distance to support points is estimated given only limited information about the data distribution, it must be very conservative to ensure correctness. Unfortunately, this inevitably leads to prohibitively high data duplication as shown above. Designing a strategy that safely bounds the support set for each partition while introducing minimum data duplication thus represents an open problem.
Second, load balancing across the partitions is critical, since a single long-running analytics task will determine the response time of the entire job. Furthermore, if the workload assigned to one machine exceeds its accommodation capacity, it will cause job failures. In distributed kNN search, the workload of each machine is determined not only by the cardinality of each core data partition, but also the size of its associated support set. The strongly interdependent nature of the partitioning and support point estimation problems complicates load balancing. On the one hand, to ensure load balance the partitioning has to take the support points into consideration. On the other hand, the support points required for a partition are determined by the distribution of data within. This raises the proverbial chicken and egg problem.
Proposed Approach. In this work we propose a distributed method that for the first time scales exact kNN search to billion point datasets. Our approach fully exploits a pivot-based partitioning strategy, so called Pivot-Based k NN Search (PkNN).
PkNN overturns the common understanding that a distributed analytics task should be completed in as few rounds as possible. We show that decomposing the kNN search into multiple rounds reveals opportunities to solve the problems of generating balanced partitions and accurately predicting support points. The rich knowledge learned in the kNN search itself can be fully explored to derive a data-driven bound on the support set much tighter than bounds based on worst-case estimation. In particular PkNN features a multi-granularity pruning strategy to accurately yet efficiently locate support points. By leveraging the learned knowledge it is able to identify and prune the set of partitions that cannot possibly contain any support points. Then at a finer individual data point granularity it introduces the concept of a boundary hyperplane to quickly discover support points based on their position relative to the hyperplane.
The support set bound and learned knowledge together are then leveraged to dynamically adjust the imperfect initial partitioning to ensure load balancing for the next round of kNN search. Our key observation here is that severe load imbalance is caused by a few far-located points in the dataset, so called outliers. By dynamically re-distributing these points, our outlier-driven partition adjustment mechanism ensures that no partition will overwhelm the resources of its assigned machine. Furthermore, it is shown to tighten the support point bound – minimizing data duplication to a rate close to 1.
Contributions. The key contributions include:
-
1.
We present PkNN, the first distributed solution that scales kNN search to billion point datasets.
-
2.
Our multiple round kNN search strategy successfully minimizes the data duplication rate and balances the workload across different machines.
-
3.
Our outlier-driven partition adjustment strategy overcomes the outlier problem in kNN search, adapting partitioning to the data distribution and available computing resources of each machine.
-
4.
Experimental evaluation on large real-world datasets shows that PkNN significantly outperforms the state-of-the-art in data duplication rate and runtime.
2 Problem Statement
Here we formalize the distributed kNN search problem. Frequently used symbols are listed in Table 1. Given a multi-dimensional dataset D, the task is to find for each point p in D a set of k other points which are most similar to p. We assume this similarity measure to be a well defined distance metric \(|\mathord {\cdot }|\) such as any \(L^p ~norm\).
Definition 1
\(k\! {\varvec{NN}}\) Search. \(\forall \) points p \(\in D\), find the set of points kNN(p) where \(kNN(p) = \{q_1, q_2, ... , q_k \in D ~|~ \forall r \in D \setminus kNN(p), |p, q_i| \le |p, r|, ~r \ne p \}\).
The naïve solution for centralized kNN search is to simply compare each point with every other point in the dataset and choose the k closest. This requires access to all points in the dataset, i.e. it is quadratic. For today’s huge datasets, the time to complete this exhaustive search is prohibitive, and furthermore, the data may be too large to be accommodated on a single machine. Therefore the kNN search task must be completed in a distributed fashion without local access to the entire dataset. We target shared-nothing platforms such as Hadoop [22] and Spark [25] where unrestricted pairwise exchange of data is not allowed between machines. Therefore our goal is to complete the kNN search autonomously for the portion of the dataset processed on each machine. To meet stringent response time requirements of big data applications, this distributed approach must effectively minimize end-to-end execution time.
Definition 2
Distributed \(k\!{\varvec{NN}}\) Search. Given a dataset D stored across n machines in a distributed compute infrastructure, perform kNN Search by processing n subsets (called cells) \(C_i\) such that \( C_1 \cup C_2 \cup ... \cup C_n \) = D on separate machines independently in parallel while minimizing end-to-end execution time.
2.1 Data Partitioning
Data partitioning is the first step of any distributed analytics task. If data is distributed among machines randomly, then it is likely that the neighbors of a given point will be sent to a different machine. Therefore a data partitioning strategy which preserves data proximity is desired. Here we adopt pivot-based data partitioning. First a small set of n initial points, or pivots, is chosen and then all data points are grouped based on their distances to the pivots. The result is a data partitioning known as a Voronoi Diagram which determines a unique division of the metric space into Voronoi cells.
Definition 3
Voronoi Cell. Given a dataset D and set of n pivots \(P = \{ v_1, v_2, \ldots , v_n\}\) we have n corresponding Voronoi cells \(\{V_i \mid V_1 \cup V_2 \cup ... \cup V_n = D, V_i \cap V_j = \emptyset \}\) and \(\forall p \in V_i, distance(p, v_i) \le distance(p, v_j), i \ne j.\)
Although pivot-based partitioning preserves the locality of the data, for points that lie along the boundaries of cells, their neighbors may still be sent to other machines. The use of a support set for each data partition ensures that the neighbors of all points in a partition can be found locally. Each cell is augmented by additional points which may be neighbors of those points near the boundary of the cell [5, 15, 24]. The points inside each cell \(C_i\) are denoted as \( C_i.core =\{p~|~p \in C_i\} \). The support set of the cell, denoted \(C_i.support\), must be sufficient to guarantee that the kNN of all core points in each cell \(C_i\) can be found among \(C_i.core\) and \(C_i.support\).
Definition 4
Support Set. The support set of a cell \(C_i\) contains at least all data points which satisfy the following two conditions: (1) \(\forall q \in C_i.support, q \not \in C_i.core \), and (2) there exists at least one point \(p \in C_i.core\) such that \(q \in kNN(p)\).
This suppport set strategy categorizes data points into two classes, namely core points and support points. Each data point will be assigned as a core point to exactly one cell, and possibly many support sets. Support points must be duplicated and transmitted multiple times. A large number of support points increases the computation costs per partition since many more points must be searched. Worst yet, it also introduces heavy communication costs which often dominate the cost of distributed methods [2]. We model these costs by the duplication rate in Definition 5. To minimize the duplication rate, the support set should contain as few points as possible.
Definition 5
Given a dataset D and a distributed algorithm A for computing the kNN of all points in D, the duplication rate \( dr(D,A)=\frac{\mid Rec(D,A)\mid }{\mid D \mid } \), where \( \mid D\mid \) represents the cardinality of D and \( \mid Rec(D,A)\mid \) the number of data records produced by the partitioning of Algorithm A.
3 PkNN: Pivot-Based Distributed kNN Search
3.1 The Overall PkNN Approach
Our Pivot-based Distributed kNN Search approach or in short PkNN not only ensures autonomous kNN computation for each data partition, but also minimizes the data duplication caused by the use of support sets. PkNN is based on our critical observation about the data dependent property of k NN distance A common understanding in the distributed analytics literature is that in shared-nothing distributed infrastructures an analytics task should be completed in as few rounds (or MapReduce jobs) as possible – ideally in one round. This avoids reading and writing the data many times, as well as high communication costs incurred from repeated shuffling phases, which are often the dominant costs of a distributed algorithm [21]. Although widely adopted in the literature, this approach does not hold in the distributed kNN search context due to the data dependent property of kNN distance. In order to complete the kNN search in one round, the distance to the support points has to be estimated a priori. However, the distance from each point to its kNN may vary considerably across a dataset. It may be small in dense areas, while very large in sparse areas. Therefore it is difficult to predict the distance to support points and in turn accurately estimate the support set of each data partition without precisely knowing the underlying data distribution characteristics. Given only limited information about the data distribution, estimates must be very conservative to ensure correctness [15]. This inevitably leads to prohibitively high data duplication mitigating the benefit of the one round solution.
The Multi-round Approach. Inspired by the data dependent property observation, PkNN no longer aims to complete the kNN search in one round. Instead it decomposes the kNN search process into two steps, namely core k NN search and support k NN search. The core kNN search step performs an initial kNN search only over the core points in each cell. The kNN produced for a point p in this step is called the core kNN of p. The key insight is that by examining the points in each cell directly we learn essential information about the characteristics of the underlying data in each Voronoi cell \(V_i\). Specifically, we determine the core-distance and support-distance of each cell to derive a tight upper bound on the distance from any core point to its support points.
After the support set of each Voronoi cell is determined, the support kNN search step then completes the kNN search over only the support points of each cell. We observe that given a core point \(p \in V_i\), its true kNN are guaranteed to be located among its core kNN and \(V_i.support\). In other words, no repeated computation ensues in our multi-round kNN search strategy. Next we detail how to utilize the core-distance and support-distance bounds to locate the support set.
3.2 Support Set Discovery
Our support set discovery method features a multi-granularity pruning strategy to accurately yet efficiently locate support points. Given a cell \(V_i\), by utilizing the support-distance \((V_i)\) and the distances between the pivots, it is able to quickly identify and prune the set of Voronoi cells \(V_j\) that do not have any chance to contain the support points of \(V_i\). Furthermore, at a finer individual data point granularity, by introducing the concept of boundary hyperplane, it quickly prunes the points based on their distances from the hyperplane and the core-distance \((V_i)\). First, we formally define the concepts of core-distance and support-distance.
Definition 6
The core-distance of a Voronoi cell \(V_i\). core-distance\((V_i) = max( |p,q| )~ \forall ~p,q \in V_i.core\) where \(q \in kNN(p) \in V_i.core\).
The core-distance of a given Voronoi cell \(V_i\) represents the maximum distance from a core point p to its kth nearest core neighbor q. The core-distance effectively defines an upper bound on the distance between any core point of \(V_i\) and the possible support points. In other words, given a point q outside \(V_i\), it is guaranteed not to be a support point of \(V_i\) if its distance to any core point of \(V_i\) is larger than the core-distance \((V_i)\).
The support-distance takes the pivot \(v_i\) of cell \(V_i\) into consideration. It captures the maximum distance of a possible support point of \(V_i\) to the pivot of \(V_i\).
Definition 7
The support-distance of a Voronoi cell. support-distance\((V_i) = max(|v_i, p | + |p, q|)\) where q is the kth nearest neighbor of \(p ~\forall p,q \in V_i.core\).
Cell Granularity Pruning. For each Voronoi cell \(V_i\), we first utilize the support-distance \((V_i)\) to determine a set of candidate support cells, as shown in Fig. 2. This is equivalent to pruning the cells which could not possibly contain support points for \(V_i\), as stated in Lemma 1.
Lemma 1
Cell Granularity Pruning. Given Voronoi cells \(V_i, V_j\) and their corresponding pivots \(v_i, v_j ~i\ne j\), if the support-distance \((V_i) \le |v_i, v_j|/2\), then \(V_j\) does not contain any support points of \(V_i\).
Proof
Recall that by Definition 3, since q is in \(V_j\), it is closer to the pivot \(v_j\) than to any other pivot. Therefore, \(|q, v_i| \ge |v_i, v_j|/2\). We give a proof by contradiction: Let some point \(q \in V_j\) be a necessary support point of cell \(V_i\), i.e., \(q \in k\textit{NN}(p)\) for \(p \in V_i\). Then by Definition 4, \(| p, q | < | p, r |\) where \(r \in V_i.core\) is the kth nearest neighbor of p out of all the core points in \(V_i\).
Assume that Theorem 1 is not true. Then we have:
This results in a contradiction. If \(|q, v_i| < |v_i, v_j|/2\), then \(q \in V_i\), which violates the original assumption.
Based on Lemma 1, we can quickly determine whether a cell \(V_j\) is a support cell of \(V_i\) by only computing the distance between their pivots. Performing this pruning at the cell level, we not only avoid unnecessary data duplication, but also reduce the number of cells each point must be checked against when mapping points to support sets.
Point Granularity Pruning. Even if a cell \(V_j\) is determined to be a support cell of \(V_i\), not every point in \(V_j\) is a necessary support point of \(V_i\). Our point granularity pruning strategy is based on the concept of a boundary hyperplane. Given two adjacent Voronoi cells \(V_i\) and \(V_j\), the boundary hyperplane is a hyperplane \(H_{ij}\) that contains the midpoint of \(V_i\) and \(V_j\). Given a point q in \(V_j\), the distance from q to \(H_{ij}\) – denoted as hp-distance \((q, H_{ij})\) (simplified as hp-distance(q) when no ambiguity arises) – and the core-distance of \(V_i\) together can determine whether q is a support point of \(V_i\).
Lemma 2
Point Granularity Pruning. Given any point \(p \in V_i, ~q \in V_j, ~i\ne j\), if hp-distance \((q) \ge \textit{core-distance}(V_i)\), then \(q \not \in k\text {NN}(p)\)
Proof
Let \(r \in V_i\) be the furthest core kNN of \(p \in V_i\). By Definition 6, \(|p,r| \le \textit{core-distance}(V_i)\). Since \(|q,p|>\) hp-distance(q) and hp-distance \((q) \ge \textit{core-distance}(V_i)\), we get \(|q,p| > \textit{core-distance}(V_i)\). Therefore, \(|q,p| > |p,r|\). This proves that q is guaranteed to be not a kNN of p.
Figures 2 and 3 illustrate the intuition behind these core and support bounds in a 2D space. In Fig. 2 the support-distance of a cell \(V_i\) is used to determine a set of shaded candidate cells which may possibly contain support points. Then in Fig. 3 the core-distance \((V_i)\) is determined by the max distance from a point to its kth nearest neighbor – in this case |p, r|. Points in the candidate support cells which fall within this distance of the linear boundaries of \(V_i\) are assigned to \(V_i.support\).
hp-distance Computation. When the commonly employed Euclidean or Mahalanobis distances are used to define the space over which the Voronoi Diagram is constructed, the boundaries of Voronoi cells are comprised of piecewise linear hyperplanes described by Theorem 1. In this case, the exact hp-distance between a point and a cell can be computed [19].
Theorem 1
The boundary between two adjacent Voronoi cells \(V_i\) and \(V_j\) is a hyperplane \(H_{ij}\) containing their midpoint. In \(\mathbf {R}^d\) the hyperplane is given as \(H_{ij} = {y : y^Tn+p = 0}\) where n is the normal vector orthogonal to the plane. \(\forall y \in H_{ij}, |y, v_i| = |y, v_j|\). The distance of any point \(p \in \mathbf {R}^d\) to the plane is \(hp-distance(q) = \frac{|s^{T}n+p|}{\Vert n \Vert _2}\).
In arbitrary metric spaces the exact computation of the hp-distance may not be straightforward. Fortunately, the lower bound on hp-distance(q) in Theorem 2 is shown in [13] via triangle inequality to hold. This lower bound can be used in place of the exact value of hp-distance(q) in Lemma 2. The proof of this conclusion is straightforward. If the lower bound of hp-distance(q) is larger than core-distance(\(V_i\)), then the exact hp-distance(q) is guaranteed to be larger than core-distance \((V_i)\).
Theorem 2
Given two cells \(V_i\) and \(V_j\) with corresponding pivots \(v_i\) and \(v_j\) and a point \(q \in V_j\), hp-distance \((q) \ge \frac{|q,v_i| - |q,v_j|}{2} \).
Point-granularity support assignment along with the cell-based pruning strategy introduced in Lemma 1 together ensure that PkNN quickly discovers the support set of each cell \(V_i\), while minimizing the data duplication rate by only including a small set of potential neighbors.
4 Outlier Driven Partition Adjustment
Our multi-round PkNN approach reduces the duplication rate dramatically and significantly outperforms the alternatives in both end-to-end execution time and scalability to the size of the dataset as confirmed in our experiments. However, our experimental study and theoretical analysis demonstrate that it still cannot handle datasets with billion point cardinality due to load imbalance. It is well known that load imbalance causes job failures if the workload assigned to one machine exceeds its processing capacity.
Support-Aware Load Balancing. Intuitively, pivot-based partitioning tends to generate partitions with a balanced workload if pivots are uniformly randomly sampled from the data utilizing for example a reservoir sampling technique [23]. More pivots are selected from dense areas and fewer from sparse areas, naturally reflecting the distribution of the data and tending to evenly partition even data with skewed distribution. Unfortunately, the workload of each machine is not only determined by the cardinality of each data partition, but also by the size of its support set. This makes the pivot-based partitioning problem and the support point estimation problem strongly interdependent. The support points are determined by the distribution characteristics of each data partition. Therefore without knowing the data partitioning it is impossible to estimate the support points.
The remedy again comes from our multi-round PkNN approach. Since PkNN decomposes kNN search into two steps, core kNN search and support kNN search, it gives us the opportunity to adjust the partitions formed at the core kNN search step and avoid an unbalanced workload during the support kNN search caused by the addition of support points. In other words, it is not necessary to solve the ‘mission impossible’ of producing partitions with perfect balanced workload before conducting the kNN search. First, uniform sampling is sufficient to produce partitions balanced enough for core kNN search. Next, based on the support set discovered via the method introduced in Sect. 3.2, we adjust partitions that could potentially overload the compute nodes.
Outlier Driven Partition Adjustment (ODA). Our partitioning adjustment method is based on the key observation that given a data partition \(V_i\), some particular points in \(V_i\) lead to a large number of support points and in turn cause severe load imbalance. These particular points, called outliers, are points that have large distance from their kth nearest core neighbor. By Lemma 2, a point q is assigned to \(V_i.support\) if the distance from q to the boundary hyperplane is smaller than the core-distance \((V_i)\). Therefore the larger the core-distance \((V_i)\), the more points \(V_i.support\) contains. Since the core-distance \((V_i)\) is determined by the point p in \(V_i\) that is furthest from its core kNN, the outliers will significantly increase the number of the support points. Leveraging this observation, our outlier driven partition adjustment, or in short ODA not only ensures no machine will be overloaded, but also minimizes the data duplication rate close to 1.
4.1 ODA: Outlier Driven Partitioning Adjustment
Definition 8
Given a compute cluster \(\mathbb {C}\) with a constraint m on the number of points that can be processed by a single machine \(M_i\) in \(\mathbb {C}\) (mapped from the resource constraint of M such as memory), the goal of ODA is to ensure that each cell \(V_i\) of dataset D satisfies the constraint: \( |V_i.core| + |V_i.support| <= m , \forall ~V_i \in D \).
Outlier Discovery. ODA first discovers outliers by translating the data cardinality constraint m put on each single machine \(M_i\) to a distance constraint \(\alpha _i\) based on the distribution of the core points in cell \(V_i\). A point o is considered as an outlier if the distance to its kth core neighbor is larger than \(\alpha _i\).
ODA first maps the irregular shaped Voronoi cell \(V_i\) generated at core kNN search step into regular hypersphere centered on the pivot \(v_i\). Assuming the core points \(V_i\).core are uniformly distributed in this hypersphere, then the radius r of \(V_i\) can be represented utilizing the mean distance \(|v_i,p| ~\forall p \in V_i.core\). The number of core points c assigned to \(V_i\) can be naturally considered as the “mass” of \(V_i\). Then the “density” of \(V_i\) can be modeled as \( density(V_i) = \frac{c}{Volume(V_i)} \). Based on the well known mathematical principle that the “volume” of a hypersphere in n dimensional Euclidean space, is proportional to the nth power of the radius r by a constant factor, \( Volume(V_i) \) can be represented by \( x \times r^n \), therefore \( density(V_i) = \frac{c}{x \times r^n} \).
Assume that the distribution of the data close to a Voronoi cell \(V_i\) will be similar to that contained within the cell, then a hypersphere \(\hat{V}_i\) centered also on the pivot \(v_i\) but with a larger radius will have the similar density to \(V_i\). Suppose \(\hat{V}_i\) contains m points, where m represent the data cardinality constraint of machine \(M_i\). Then we have
Here \(\alpha _i\) denotes the allowable distance to expand \(V_i\) to a sphere \(\hat{V}_i\) that contains no more than m points. This value \(\alpha _i\) can be computed as follows.
For each Voronoi cell \(V_i\), \(\alpha _i\) gives a customized max threshold on the distance to points in \(V_i\).support subject to resource constraints and the data distribution of \(V_i\).core.
Lemma 3
If \(|p,q| < \alpha _i~ \forall ~p,q \in V_i.core\) where \(q \in kNN(p) \in V_i.core\),
then \(|V_i.core| + |V_i.support| < m\).
Proof
If the core k-distance of \(\forall \) point p \(\in \) \(V_i\).core is smaller than \(\alpha _i\), then all points in \(V_i\).support are covered in hypersphere \(\hat{V}_i\). Since \(\hat{V}_i\) covers at most m points, this proves \( |V_i.core| + |V_i.support| < m \).
Based on Lemma 3, points \(p \in V_i.support\) are identified as outliers of cell \(V_i\) if their distance to their core kNN cause core-distance \((V_i)\) to be larger than \(\alpha _i\).
Partition Adjustment. To ensure that outliers cannot skew a cell \(V_i\) and cause job failure, \(V_i\) will be adjusted before conducting the support kNN search step by redistributing the outliers within the compute cluster. This eliminates the influence of the outliers on our data-driven bounds, while still leaving the original partitioning intact.

The overall outlier-driven partition adjustment approach (ODA) is shown in Algorithm 1. First, for each cell \(V_i\) the information necessary to compute \(\alpha _i\), namely the average distance from each point to the pivot \(v_i\) can be easily collected during the initial pivot-based data partitioning phase (Line 2). Then outliers are identified during core kNN search. After the core k-distance of a point p is calculated, p is marked as an outlier if its core k-distance exceeds the threshold \(\alpha _i\) (Line 3). Next when determining the support set of \(V_i\), these outliers are treated as additional “special” pivots. These special pivots differ from regular pivots in that no additional core points are assigned to their partitions (Line 4). Since an outlier o would have already found their kth nearest neighbor from among the core points (core kNN) in their original partition \(V_i\), the distance from o to its kth core kNN determines the core-distance of its new special partition. This in turn is utilized to determine the support points of o (Line 5). In the support kNN search step, kNN search is conducted only for the single core outlier point of each of these “special” partitions (Line 6). Since by their nature the outliers are far from all other points, it is unlikely that many additional support points will be mapped to the newly formed special partitions.
By discovering and handling the outliers in this way, ODA ensures that no partition \(V_i\) will exceed the processing capacity of each machine. Furthermore, the overall data duplication rate of the whole dataset is further reduced to be close to 1 as confirmed in our experiments. The overall structure of the full-fledged PkNN framework is shown in Fig. 4.
5 Experimental Evaluation
5.1 Experimental Setup
All experiments are conducted on a shared-nothing cluster with one master node and 28 slave nodes. Each node consists of 16 core AMD 3.0 GHz processors, 32 GB RAM, 250 GB disk, and nodes are interconnected with 1Gbps Ethernet. Each server runs CentOS Linux (kernel version 2.6.32), Java 1.7, Hadoop 2.4.1. Each node is configured to run up to 4 map and 4 reduce tasks concurrently. Speculative execution is disabled to boost performance. The replication factor is set to 3.
Datasets. We evaluate PkNN on real world data using OpenStreetMap [1], one of the largest real datasets publicly available, which has been used in similar research work [26]. It contains geolocation information for physical landscape features such as a buildings and roads all over the world. Two attributes are used, longitude and latitude. Hierarchical datasets (Shown in Table 2) evaluate the scalability of PkNN with regard to the data size. The datasets grow from a Massachusetts dataset of 10 million points to over 1 billion points covering more than the western hemisphere.
Real data are also used to evaluate the performance of PkNN in higher dimensions. The Sloan Digital Sky Survey (SDSS) [8] is one of the largest astronomical catalogs publicly accessible. We extracted a dataset containing 100 million records from the thirteenth release of SDSS data. In this experiment we utilize seven numerical attributes including Right Ascension, Declination, three Unit Vectors, Galactic longitude and Galactic latitude. The TIGER [9] dataset contains 70 million line segments, representing GIS features of the United States. Four numerical attributes giving the longitude and latitude of two endpoints of line segments are used.
Methods. We compare PkNNFootnote 1 against two state-of-the-art distributed solutions for kNN search in MapReduce. The first called PBJ [15] uses pivot-based partitioning to perform kNN Join. The authors of the [15] generously shared their code, which we adapted to run on a single dataset. The second called Spitfire [5] uses grid-based partitioning to perform kNN search on a message-passing architecture. We have implemented their method and adapted it for MapReduce.
Metrics. End-to-end execution time is measured, which is common for the evaluation of distributed algorithms. Furthermore, the execution time for the key stages of the MapReduce workflow is broken down to evaluate the performance of different stages of computation. The other key metric is the duplication rate (Definition 5). This measure, calculated as the total number of core and support points processed divided by the number of points in the input dataset, captures how effectively supporting areas are bounded.
5.2 Evaluating Data Duplication Rates
We first evaluate the data duplication rate of the three methods with respect to the number of data partitions. For a truly scalable solution, the benefit of adding partitions (necessary as the data size grows) should not be outweighed by the increased communication costs. Experiments are conducted on the Massachusetts dataset containing 10 million records. This data set has areas of varying density throughout the domain space. k is fixed at 5 and the number of pivots is varied from 50 to 1100.
To perform a fair comparison of PkNN and PBJ, they are required to use the same pivot set, as the choice of pivots impacts performance significantly. Therefore we omit the grouping step introduced in [15], which starts with a very large number of pivots and then groups them into a small number of final partitions. As their own evaluation shows, this grouping step only results in a modest decrease in execution time [15]. The number of partitions in Spitfire is tuned to a similar amount, although the equi-depth partitioning algorithm [5] does not allow us to set an exact number of partitions. Both the end-to-end runtime as well as the data duplication rate are measured for all methods.
Figure 5a shows PkNN and Spitfire clearly attain much lower data duplication than PBJ (shown on the left), and consequently execution time (shown on the right). Even using a small number of pivots, the minimum duplication rate for PBJ is around 30. PkNN and Spitfire on the other hand remain close to the optimal rate of 1. The PBJ method shows a quadratic increase in running time. Figure 5b compares the overall execution time for PkNN and Spitfire. For these methods, adding more data partitions improves runtime. Both methods perform similarly on this small data size. However, as we demonstrate in Sect. 5.3, PkNN outperforms Spitfire due to its computational complexity as the data size increases, despite both methods achieving low data duplication rates.
5.3 Evaluating Scalability
In this section the scalability of PBJ, Spitfire, and PkNN with regard to the data size is evaluated. For this set of experiments we use 2d data from the Open Street Map dataset and fix k at 5. Figure 6 shows the end-to-end execution time of each method on datasets of varying size, with the time for each key stage of the MapReduce workflow broken down. In Fig. 6a we observe that the single round kNN search approach of the PBJ method is almost 20 times slower than the two kNN search rounds in PkNN. Attempts to evaluate PBJ on data larger than 10 million points were not successful due to job failures caused by the high data duplication rate.
Figure 6b and c show PkNN outperforms Spitfire by a factor of 3 and a factor of 7 respectively. The first kNN job in Spitfire performs the support discovery step using the concept of point hiding [5]. These experiments clearly show the computational complexity of this approach hinders performance when the data contains hundreds of millions of points. Figure 6d evaluates the performance of PkNN on a billion point dataset, which neither competing method could handle. This demonstrates that PkNN truly scales to handle modern data quantities.
Table 2 shows the dataset sizes and parameters used for evaluation. To choose the number of pivots, it was observed that a ratio of 1: 30,000 points performed well on the small dataset in initial experiments. Fewer pivots were experimentally determined to perform better on larger data. Attempts to run PkNN on datasets larger than 10 million without the ODA partition adjustment mechanism lead to job failures. However, by identifying a small fraction of the data as outliers in each dataset ODA achieves a more balanced workload allowing for the processing of these large datasets, as well as data duplication rates close to 1.
5.4 Evaluating the Impact of the Number of Dimensions
Finally we show that PkNN effectively handles multidimensional data at scale. The PBJ method cannot accommodate data beyond the 10 million points, and Spitfire only supports 2-dimensional data. Table 3 shows the performance of PkNN on the Tiger and SDSS datasets which contain 70 and 100 million records and four and seven dimensions respectively. We can see PkNN continues to achieve low data duplication rates under 2 on real multidimensional data as well as fast runtime.
6 Related Work
kNN search is a well-studied problem with many solutions to mitigate its high computational complexity (O(\(n^2\))). In centralized algorithms, spatial indexing structures are typically used, such as grid-based indices for low dimensional data or hierarchical tree structures [6, 11, 14], as they achieve an O(nlogn) expected complexity.
In recent years, a number of distributed solutions for kNN search have been proposed for message-passing and peer-to-peer distributed systems [5, 12, 18]. These approaches cannot be utilized on modern distributed architectures such as MapReduce [22] and Spark [25] which are the focus of our work due to their scalability, fault tolerance, and ability to run on commodity hardware. A recent methods for kNN classification have targeted these platforms [16, 17], however they utilize a broadcast join technique, where the neighbors of a small test set of points are found among a larger training set distributed across a compute cluster. This technique is not applicable to our problem where we aim to find all neighbors of all points in large datasets (over 1 billion points) which cannot be processed in their entirety on a single machine.
A MapReduce based kNN search approach was proposed in [15]. Similar to PkNN this approach also utilizes pivot-based partitioning to divide the dataset. Worst-case estimation based on the distance from pivots to a small number of points in each partition bounds the size of the supporting sets. As shown in their experiments and confirmed by our results (Sect. 5.2), this leads to extremely high data duplication (upwards of 20x the size of the original dataset). Although an adaptation of the bound and pivot selection method is presented in [24] as part of system specific to spatial data, the worst case estimation of support sets still cannot scale to large datasets due to the duplication rate. Our multi-round PkNN approach effectively cuts the duplicate rate close to be 1 by adopting a multi-round strategy.
The Spitfire approach [5] effectively improves over the duplication rate of [15, 24] by utilizing grid-based partitioning to divide the data and bound the support set. However, its point hiding concept to discover support sets leads to high computation costs as the data size increases, as confirmed by our experiments (Sect. 5.3). Spitfire is also specific to two-dimensional geo-location datasets while our PkNN approach is effective in supporting the general case of multi-dimensional datasets.
7 Conclusion
This work presents PkNN, an innovative distributed approach for kNN search over modern distributed architectures. We introduce a multi-round computation strategy along with data-driven bounds and a tiered support discovery technique which effectively limit data duplication. An outlier-driven partition adjustment mechanism ensures load balance. PkNN for the first time scales kNN search to billion-point real world datasets and gracefully handles multidimensional data.
Notes
- 1.
PkNN source code available at http://solar-10.wpi.edu/cakuhlman/PkNN.
References
Open Street Map. http://www.openstreetmap.org/. Accessed 23 Apr 2016
Afrati, F.N., Sarma, A.D., Salihoglu, S., Ullman, J.D.: Upper and lower bounds on the cost of a map-reduce computation. Proc. VLDB Endow. 6(4), 277–288 (2013)
Ankerst, M., Breunig, M.M., Kriegel, H.P., Sander, J.: OPTICS: ordering points to identify the clustering structure, pp. 49–60. ACM Press (1999)
Breunig, M.M., Kriegel, H.P., Ng, R.T., Sander, J.: LOF: identifying density-based local outliers. In: ACM SIGMOD International Conference on Management of Data, pp. 93–104. ACM, New York (2000)
Chatzimilioudis, G., Costa, C., Zeinalipour-Yazti, D., Lee, W.C., Pitoura, E.: Distributed in-memory processing of all k nearest neighbor queries. IEEE Trans. Knowl. Data Eng. 28(4), 925–938 (2016)
Ciaccia, P., Patella, M., Zezula, P.: M-tree: an efficient access method for similarity search in metric spaces. In: Proceedings of 23rd International Conference on Very Large Data Bases, pp. 426–435 (1997)
Cover, T., Hart, P.: Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 13(1), 21–27 (1967)
Dawson, K.S.: The SDSS-IV extended baryon oscillation spectroscopic survey: overview and early data. Astron. J. 151(2), 44 (2016). http://stacks.iop.org/1538-3881/151/i=2/a=44
Eldawy, A., Mokbel, M.F.: SpatialHadoop: a MapReduce framework for spatial data. In: International Conference on Data Engineering, pp. 1352–1363. IEEE (2015)
Ester, M., Kriegel, H.P., Sander, J., Xu, X.: A density-based algorithm for discovering clusters in large spatial databases with noise. In: KDD, vol. 96, pp. 226–231 (1996)
Guttman, A.: R-trees: a dynamic index structure for spatial searching, vol. 14. ACM (1984)
Haghani, P., Michel, S., Cudré-Mauroux, P., Aberer, K.: LSH at large-distributed KNN search in high dimensions. In: International Workshop on the Web and Databases (2008)
Hjaltason, G.R., Samet, H.: Index-driven similarity search in metric spaces (survey article). ACM Trans. Database Syst. 28(4), 517–580 (2003)
Lin, K.I., Jagadish, H.V., Faloutsos, C.: The TV-tree: an index structure for high-dimensional data. Int. J. Very Large Data Bases 3(4), 517–542 (1994)
Lu, W., Shen, Y., Chen, S., Ooi, B.C.: Efficient processing of k nearest neighbor joins using mapreduce. Proc. VLDB Endow. 5(10), 1016–1027 (2012)
Maillo, J., Ramírez, S., Triguero, I., Herrera, F.: kNN-IS: an iterative spark-based design of the k-nearest neighbors classifier for big data. Knowl.-Based Syst. 117, 3–15 (2017)
Maillo, J., Triguero, I., Herrera, F.: A mapreduce-based k-nearest neighbor approach for big data classification. In: Trustcom/BigDataSE/ISPA, vol. 2, pp. 167–172. IEEE (2015)
Novak, D., Zezula, P.: M-Chord: a scalable distributed similarity search structure. In: Proceedings of the 1st International Conference on Scalable Information Systems, p. 19. ACM (2006)
Ramaswamy, S., Rose, K.: Adaptive cluster distance bounding for high-dimensional indexing. IEEE Trans. Knowl. Data Eng. 23(6), 815–830 (2011)
Ramaswamy, S., Rastogi, R., Shim, K.: Efficient algorithms for mining outliers from large data sets, vol. 29, pp. 427–438. ACM (2000)
Sarma, A.D., Afrati, F.N., Salihoglu, S., Ullman, J.D.: Upper and lower bounds on the cost of a map-reduce computation. In: Proceedings of the VLDB Endowment, vol. 6, pp. 277–288. VLDB Endowment (2013)
Shvachko, K., Kuang, H., Radia, S., Chansler, R.: The hadoop distributed file system. In: IEEE 26th Symposium on Mass Storage Systems and Technologies, pp. 1–10. IEEE (2010)
Vitter, J.S.: Random sampling with a reservoir. ACM Trans. Math. Softw. 11(1), 37–57 (1985)
Xie, D., Li, F., Yao, B., Li, G., Zhou, L., Guo, M.: Simba: efficient in-memory spatial analytics. In: ACM SIGMOD International Conference on Management of Data, pp. 1071–1085 (2016)
Zaharia, M., Chowdhury, M., Franklin, M.J., Shenker, S., Stoica, I.: Spark: cluster computing with working sets. HotCloud 10, 10 (2010)
Zhang, C., Li, F., Jestes, J.: Efficient parallel KNN joins for large data in mapreduce. In: Proceedings of the 15th International Conference on Extending Database Technology, pp. 38–49. ACM (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Kuhlman, C., Yan, Y., Cao, L., Rundensteiner, E. (2017). Pivot-Based Distributed K-Nearest Neighbor Mining. In: Ceci, M., Hollmén, J., Todorovski, L., Vens, C., Džeroski, S. (eds) Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2017. Lecture Notes in Computer Science(), vol 10535. Springer, Cham. https://doi.org/10.1007/978-3-319-71246-8_51
Download citation
DOI: https://doi.org/10.1007/978-3-319-71246-8_51
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-71245-1
Online ISBN: 978-3-319-71246-8
eBook Packages: Computer ScienceComputer Science (R0)