Abstract
Early detection and accurate diagnosis for liver disease are very important. Due to the defects inherent in the ultrasound images and the complexity appearance of diseases, automatic classification for liver diseases in ultrasound images is a challenging task. In this paper, we introduce a novel method to classify focal liver lesions in ultrasound images. At first, we use an automatic image segmentation algorithm to delineate the lesion region. Then, according to the characteristics of liver lesions, we design a new image feature which is discriminative to liver lesions. Finally, six image features are processed by an improved sparse representation classifier to identify the diseases. We expand the sparse representation dictionary to optimize the classifier. Experimental results have shown that the proposed method could improve the classification accuracy in comparison with other state-of-the-art classifiers. It should be capable of assisting the physicians for liver disease diagnosis in the clinical practice.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Liver cancer is the sixth most common cancer and the third leading cause of death in the word [1]. Early detection and accurate diagnosis for liver disease are very important, which are helpful to increase the chances for survival [2]. Currently, Ultrasound (US) imaging is the most widely spread imaging method because it is noninvasive, low cost and easy to operate.
Generally, the clinical diagnosis of liver diseases in ultrasound images is decided by specialized physicians’ visual interpretation. The diagnosis is time-consuming and depends on the physicians’ clinical experience. Hence, the demand for computer-aided diagnosis (CAD) in ultrasound images increased rapidly. CAD is able to assist the physicians and improve the diagnostic accuracy.
With the development of computer technology, pattern recognition, and machine learning methods, researchers have done some effort on computer aided diagnosis of ultrasound images. In liver diseases diagnosis, most studies focused on diffused liver diseases [3,4,5] and focal liver diseases [6, 7]. Diffused liver diseases always attach to the whole liver. Hepatic adipose infiltration and cirrhosis are the most common diffused liver diseases. Focal liver diseases are concentrated over a quite small area of the tissue and it is very difficult to identify. Therefore, focal liver diseases classification is a more challenging task. Hepatic cyst (cyst), hemangioma (Hem) and hepatocellular carcinoma (HCC) are the most common focal liver diseases.
Typically, there are three main stages for liver diseases classification systems, including selection of region-of-interests (ROI), feature extraction and diseases classification by classifier.
For diffused liver diseases, Sabih and Hussain [3] and Owjimehr et al. [4] proposed automatic classification methods to diagnose liver steatosis in levels of Normal, Fatty and Heterogeneous. Sabih and Hussain [3] chose lesion regions as region-of-interests (ROI) and then used Wavelet Packet Transform (WPT) to extract regional statistical features. They finally achieve automatic classification by using a multi-class linear Support Vector Machine (SVM). Owjimehr et al. [4] mainly extracted Completed Local Binary Pattern (CLBP) features, and used SVM to do classification. In Alivar et al. [5], the ROI was cropped manually in ultrasound images and then extracted Wavelet Packet energy features, Gabor filter energy features and Gray Level Co-occurrence Matrix (GLCM) features. At last, normal, fatty and cirrhotic liver ultrasound images were classified by using K Nearest Neighbor (KNN) classifier.
For focal liver diseases, Jeon et al. [6] proposed an approach to recognize three kinds of focal liver lesions, including cysts, hemangiomas, and malignancies. They selected multiple different ROI, and then extracted five types of feature sets, containing GLCM features, First Order Statistics (FOS) features, Algebraic Moment Invariant (AMI) features, Auto-correlation (AC) features and Laws’ texture energy measure (LTEM) features. In the end, Support Vector Machine (SVM) was used to reach the final goal. Raghesh Krishnan and Radhakrishnan [7] employed isocontour segmentation method for ROI segmentation. Then they put GLCM features and fractal features extracted from ROI into Artificial Neural Networks (ANN), achieving the classification on ten types of diffused and focal liver diseases.
In order to improve the accuracy of classification for liver lesions, most researches made efforts on extracting effective feature. Because various liver diseases have their own texture expressions in ultrasound images [8], texture feature has been widely used in classification. However, it is not enough to borrow the existing features that were designed for general image classification. We should carefully observe the characteristic of the lesions in ultrasound images to design specific image features.
On the other hand, current researches used common classifiers, such as SVM, ANN, KNN, to do classification. Most of these classifiers need training stages and large samples. However, as we know, it is hard to get plenty of medical images.
In recent years, Sparse Representation-based Classification (SRC) has been successfully used to solve problems in computer vision area, such as face recognition [9]. Different from other classifiers, the SRC is a nonparametric learning method which does not need a training process. SRC only uses the training data to construct an over-complete dictionary.
The SRC is just attracting attention in medical image recognition nowadays. Currently we find seldom papers of applications in ultrasound image, including classification for breast ultrasound image [10] or echocardiography [11]. In the work of [10, 11], researchers constructed the dictionary just by extracting the image feature vectors from training samples without considering the efficiency of features. On the one hand, in order to make the dictionary over-complete, the number of training samples should be far larger than the dimensions of features. It means that plenty of training samples are needed to construct such an over-complete dictionary. Meanwhile, the quality of dictionary may be effected by an incompact structure. The atoms of different classes in the dictionary are too similar to be partitioned, due to the intrinsic defects in ultrasound images like the speckle noise, artifacts and low contrast, and the diversity of different liver diseases. This problem will lead to false classification for liver diseases. In order to solve this problem, K-SVD based algorithm [12] is the most common method used for dictionary optimization. However, K-SVD based algorithms are computationally complex and need extra training samples because it contains complex singular value decomposition in iterative procedure.
In this paper, we propose a new method for focal liver lesion classification, including hepatic cyst (cyst), hepatic hemangioma (Hem), and hepatocellular carcinoma (HCC). Our main contributions are: (1) We use automatic image segmentation algorithm to acquire the lesion region, and select both normal region and lesion region in the ultrasound image, as our region of interests (ROI), to reduce the impact of individual differences; (2)According to the pathological characteristics of lesions and their diversity appearances in ultrasound images, we design a specific image feature named mutation ratio. Combining with GLCM and fractal features, the features in our method have a good discriminative ability for focal liver lesions classification; (3) We propose an effective dictionary optimization method named dictionary expansion based on sparse reconstruction (DESR) for SRC to classify lesions. The experimental result shows that our proposed method has an excellent performance in the focal liver lesion classification. The overall accuracy of the method is 97.76%.
This paper is organized as follows: Sect. 2 introduces the proposed method. Section 3 presents experimental results and performance analysis. Finally we draw a conclusion in Sect. 4.
2 Methods
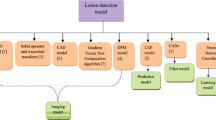
In this paper, we propose an automatic classification method of liver ultrasound images based on sparse representation. It involves multiple ROI selection, feature extraction, and classification based on sparse representation. The architecture overview is shown in Fig. 1.
2.1 Multiple ROI Selection
In this paper, we choose the lesion and normal region as two ROIs considering the contrast between the lesion and normal region.
Automatic lesion region selection.
We use an automatic energy-based region growing method (AERG) [13] of our previous work to get the lesion region automatically.
The lesion region is the focus in the ultrasound image of focal liver lesion. At present, most of the methods manually or semi-automatically select a fixation of the ROI that includes the lesion region [5, 7]. It is arduous and time-consuming. In this work, we use an automatic method to select the lesion region as ROI. The method makes the segmentation procedure timesaving and laborsaving. And the segmentation result is more accurate.
The specific steps are as follows.
-
(1)
Delineating the boundary of focal liver lesion by AERG.
At first, the seed point of lesion region is automatically selected by sparse reconstruction algorithm. Then the region growing process is controlled by a novel energy function including both internal and external energy, so as to make the edge of the region converge to the contour of the lesion accurately and keep a small internal difference at the same time. The details of this method can be found in reference [13].
-
(2)
Getting a minimum enclosing rectangle of the focal liver lesion region.
-
(3)
Cutting the rectangle area as a lesion image R1.
Figure 2(a) is the original liver ultrasound images. The first line is an example of Hem image, the second line is cyst image and the last line is HCC image. Figure 2(b) shows the segmentation results of Fig. 2(a) using AERG. The green rectangle in Fig. 2(c) is the enclosing rectangle of lesion region.
Normal region selection.
In order to describe the comparison of lesions and normal area, we also select the normal region as a ROI.
In ultrasonic diagnosis, the descriptors of echoes, such as hyperechogenicity, hypoechogenicity, echoless, echo enhancement etc., are often used for diseases identification in ultrasound image. Due to the diversity of ultrasonic equipments and manipulators, the same region may have different appearances in different equipments and manipulators. Therefore, the absolute value of a single area is not comparable in ultrasound images. The descriptors should be given under comparison between the lesion and normal region of the same image to reduce such individual differences.
We extract a rectangle region outside the lesion region as the normal region. The normal region should lie in the liver parenchyma with uniform texture and don’t contain blood vessels and bile stores. As shown in Fig. 2(c), the blue rectangle region is selected as the normal region R2.
2.2 Feature Extraction
Generally, lesion disrupts uniform and regular texture distribution of the normal liver. The texture in lesion region has heterogeneous and distinctive characteristics in the ultrasound image. Therefore, the texture features play an important role in classification of ultrasound images [14]. According to the characteristics of the lesion region, we deliberately design a new feature called mutation ratio. Along with the GLCM and fractal features, we use 6 dimensional features for the following image classification.
GLCM feature ratio.
The GLCM is a common and useful method for texture feature analysis [15, 16]. In order to eliminate the influence of different equipments or operators, we use the GLCM feature ratios of the lesion region R1 and normal region R2.
The specific method is as follows. (1) Lesion region R1 and normal region R2 are respectively computed three features (correlation, energy and homogeneity) of the GLCM at four directions [0°, 45°, 90°, 135°]. (2) Then the averages of features in four directions are computed to get the six features, namely correlation of R1(COR_R1) and R2(COR_R2) energy of R1(EN_R1) and R2(EN_R2), homogeneity of R1(Hom_R1) and R2(Hom_R2). (3) The features ratios of R1 to R2 are defined as features of GLCM, listed in formula (1):
Fractal features.
We use fractal dimension and lacunarity as the fractal features in this paper.
Fractal geometry can effectively describe the roughness of nature surface. It has been successfully applied in ultrasound images to distinguish the roughness and texture characteristics.
Fractal dimension.
Fractal dimension closely corresponds to human visual perception of roughness. It is invariant to image scale and rotation. Generally, the image surface is rougher, the fractal dimension value is bigger.
We use the differential Box-Counting approach proposed by Sarkar and Chaudhuri [17] to compute fractal dimension of image. It is calculated as:
N r is counted for different values of r.
As we can see from Fig. 2(a), the appearances of the three lesion regions are various. The cyst is composed of clear fluid and its inner structure is very simple. Therefore, the appearance cyst area is smooth with minimum fractal dimension. Hem is a vascular coil and its structure is very complex, so its appearance is rough with the maximum fractal dimension. The pathological conditions of HCC are various, so the roughness of its appearance is fluctuant.
Lacunarity.
Lacunarity is a feasible feature to measure textures in terms of gap distribution. Due to the fact that different texture may have the same or similar fractal dimension values, the fractal dimension can’t determine the image alone. The notion of lacunarity, which is related to the degree of translational invariance, makes it possible to distinguish the images with the same fractal dimension values [18].
We adopt the box-counting method [18] to compute the lacunarity and set t = 2. which is performed:
Mutation ratio.
We design a specific texture feature of mutation ratio based on the texture characteristics of different lesion regions in ultrasound images. The mutation ratio is defined to describe the density of key points, which can represent the complexity of the lesion inner structure effectively.
Those pixels that have the obvious difference with the neighboring pixels are defined as key points. Cyst is just clear fluids, whose structure is very simple. It is an echoless area in ultrasound image, so its texture is uniform and there is almost no key point. Hem which is filled with blood cells and blood vessels has large internal changes inside. Therefore, a lot of key points can be detected in Hem images. HCC appearance is mixed echogenicity and the number of key points stands between the Cyst and Hem. As shown in Fig. 2(d), the blue circles are the key points detected in the images. The key point distributions of the three images are very different.
The main stages of computing the mutation ratio are as follows. (1) Detecting key points of liver ultrasound image. We use the method in [19] to extract the key points. (2) Counting the total number φ of key points in the lesion region R1. (3) A ratio φ to the area Area of the lesion’ enclosing rectangle are defined as the mutation ratio.
Experiments in Sect. 3 will show the effectiveness of this feature in classification. Equation (4) shows the specific definition mutation ratio d:
Feature normalization.
In summary, the final feature vector \( f \) is composed of 3-dimensional GLCM feature ratios, 2-dimensional fractal features and 1-dimensional mutation ratio feature. Then a normalization is done on vector f to normalize all features to (0, 1).
2.3 The Improved Sparse Representation Classification
The core of Sparse Representation-based Classification (SRC) is that the test image can be presented as a linear combination of those training samples from the same class. If we represent the test sample in an over-complete dictionary whose base elements are all the training samples, the representation is naturally sparse, involving only a small fraction of the overall training database [9]. Different from other classifiers, SRC does not need a train process but only need the training data [11].
Sparse representation-based classification [9].
Suppose that \( D = [D\_1,D\_2, \ldots ,D\_i, \ldots ,D\_c] \) is a matrix of the entire training set with n training samples of c classes, where \( D_{i} = [f_{i1} ,f_{i2} , \ldots ,f_{{in_{i} }} ] \) is the sub-set of the training samples from class i and \( f_{ig} \) is its gth feature vector.
A new test sample y is from one of the classes in the training set. It can be expressed as a linear combination of the representation dictionary D:
where \( x = [0, \ldots ,0,x_{1}^{i} ,x_{j}^{i} , \ldots ,x_{n}^{i} ,0, \ldots ,0]^{T} \, \in \,R^{n \times 1} \) is a coefficient vector, whose coefficients are remarkable for the class the test sample belongs to, while for the other classes, the coefficients are almost equal to zeros. x is obtained by solving the linear Eq. (6):
Where \( \left\| \bullet \right\|_{1} \) is l1-norm, \( \left\| \bullet \right\|_{2} \) is l2-norm.
Ideally, if the test sample y belongs to a certain class, the coefficients in the estimated \( \hat{x} \) not within this class should all be zeros. However, noise and modeling error may lead to small nonzero entries associated with multiple object classes.
The noisy model is modified as:
where \( \varepsilon \) is a noise term.
We Define \( \delta_{i} \left( x \right) \) as a classification vector, whose entries are zeros except for those associated with the class i. Using only the coefficients associated with the ith class, we can reconstruct the given test sample y as:
We then compute the residual \( r_{i} \left( y \right) \) between y and \( \hat{y}_{i} \):
The smaller \( r_{i} \left( y \right) \) is, the more similar the true sample y and reconstructed sample \( \hat{y}_{i} \) are. Therefore, we classify y based on these reconstruction samples by assigning it to the object class that minimizes the residual \( r_{i} \left( y \right) \) between y and \( \hat{y}_{i} \):
Dictionary expansion based on sparse reconstruction.
In this section, we propose a method of Dictionary Expansion based on Sparse Reconstruction (DESR) to construct a good dictionary for SRC, and then improve the classification accuracy of SRC.
Generally, by using sparse representation method, one image can be presented as a linear combination of a small number of atoms from predefined redundant dictionary. Therefore, the good or bad of the dictionary determinates the performance of sparse representation, and influences the final classification results. Hence, it is really necessary to construct a good redundant dictionary.
Traditional SRC does not get an ideal sparse representation which leads to an unsatisfied classification result. Traditional SRC directly simply uses the samples in the training set as atoms to construct dictionary. However, in order to get a redundant dictionary, a lot of training samples should be involved to make the number of training samples far more than the dimensions of the features. Nevertheless, we can’t get so many training samples in medical image classification.
On the other hand, a good dictionary is supposed to have compact inner structure. It means that the difference between samples in the same class should be small, while be big between different class so as to make the reconstruction error small enough to classify the test samples correctly. However, due to the intrinsic defects and the complexity appearances of ultrasound image, sometimes the textures of the image appearances are very similar in different focal liver lesions. The dictionary gets an incompact structure when it is constructed by only using the samples in the training set as atoms directly.
Aiming to solve the problem in dictionary construction, we propose a method for Dictionary Expansion based on Sparse Reconstruction to construct a good dictionary.
In order to get an over-complete dictionary with good discriminative capability, we add a lot of new atoms to expand the dictionary. The dictionary is redundant and has a compact structure after expanding, which lead to a better performance in classification.
The details of DESR are shown as follows: (1) Constructing the initial dictionary \( D = [f_{1} ,f_{2} , \ldots ,f_{k} , \ldots ,f_{n} ] \) by using the entire training set with n training samples of c classes, where \( f_{k} \) is the kth training sample. (2) Taking out atom \( f_{k} \) which belongs to the ith class from dictionary D in turn. Using the rest of atoms in D to sparsely represent \( f_{k} \), and obtaining the sparse coefficients via (7). (3) Respectively using only the coefficients associated with the jth class \( \left( {j = 1,2, \ldots ,c} \right) \) to reconstruct \( f_{k} \) via (8). We can get c reconstruction samples \( \left\{ {\hat{f}_{k1} , \ldots ,\hat{f}_{kj} , \ldots ,\hat{f}_{kc} } \right\} \). (4) Computing all the residuals between \( f_{k} \) and each class-based reconstruction samples \( \left\{ {\hat{f}_{k1} , \ldots ,\hat{f}_{kj} , \ldots ,\hat{f}_{kc} } \right\} \). We can find the reconstruction sample \( \hat{f}_{kj} \) which corresponds to the minimum residual. If j is equal to i (namely, reconstruction sample is in the same class with original sample) and the minimum residual is less than a predefined value, we think the reconstruction sample \( \hat{f}_{kj} \) is small within-class scatter but big between-class scatter. Then, the reconstruction sample \( \hat{f}_{kj} \) will be added to the original dictionary D to improve redundant and discriminative capability (Table 1)
.
Classification of focal liver lesions.
We randomly select a certain number of samples as training samples and the rest as testing samples in the dataset. Training samples are used to construct the dictionary. And testing samples are classified through spare representation.
Figure 3 indicates the residual errors of a cyst and HCC ultrasound image which is correctly classified by our method and misclassified by traditional SRC.
As shown in Fig. 3, by using our method, the smallest residual error of a test image lies in the correct class. However, by using traditional SRC method, the smallest residual error of a test image lies in the incorrect class. It leads to the incorrect classification for this sample. For example, the smallest residual error of the cyst image lies in the cyst class in our method, while lies in HCC in traditional SRC method.
The above analysis shows that SRC with DESR is more effective, and get better classification performance than the traditional SRC dictionary.
3 Experimental Results and Comparison
In this work, the dataset is comprised of 108 ultrasound image including 36 HCC, 36 Hem and 36 Cysts. All these liver lesions’ categories were identified through the clinical pathological detection.
We employ leave-one-out cross validations in experiments. That is to say, we do the experiments with a training set formed from all but one case of the total data set. The single remaining sample is tested and compared with its labeled class. This process has been repeated 108 times until all samples were tested.
We extract GLCM features, fractal features and mutation ratio to make up six dimensional features, and then accomplish the Sparse Representation-based Classification with DESR to classify the three common focal liver diseases.
Five indices based on the true positive (TP), the false positive (FP), the true negative (TN), and the false positive (FP) are used to evaluate the performance of classifiers, which are the overall accuracy (ACC), the sensitivity (SEN), the specificity (SPE), the positive predicative value (PPF), and the negative predictive value (NPV) [20].
Table 2 presents the classification results on three types of focal liver lesions by our method. The classification accuracy is up to 97.76% and the rest indices are also higher than 97%, which means our algorithm is indeed effective to classify focal liver diseases.
Figure 4 lists some examples correctly classified by the proposed method.
3.1 Comparison of Features Subsets
In order to further validate the effectiveness of our features, we compare the performance of three different feature sets by using SRC of DESR. The results are shown in Table 3.
Table 3 illustrates the following conclusion. (1) Compared with the feature set including only GLCM features and fractal features, the performance is greatly enhanced after adding feature of mutation ratio. It proves the effectiveness of mutation ratio feature. (2) We extract the best seven dimensional features in paper [8] to be a comparison on our dataset. The feature set contains Spatial Gray Level Co-occurrence Matrix, Fourier Power Spectrum, the Statistical Feature Matrix (SFM) texture, the Texture Energy Measures (TEM) suggested by Laws, and the fractal-based features. The results show that our features perform better and using GLCM Ratios is effectual, especially in SEN and PPV, which are the most important indexes in clinical diagnosis.
3.2 Comparison of Different Classifiers
In this section, the performance of our proposed classifier is compared with some state-of-the-art classifiers, including the traditional SRC, the SVM and the BPNN. The six features proposed in this paper is applied to all classifiers. We use the function provided by neural network tools in MATLAB for BPNN and SVM classifier provided by LIBSVM [21] to accomplish these classifiers. For BPNN, we select ten nodes as hidden layers and for SVM, we choose linear function as our kernel function.
Table 4 shows the performances of different classifiers. It shows that our method gets the best results and all indexes are better than those in other outstanding methods.
Moreover, all classifiers achieved the overall correct percent over 90. It means that the proposed six dimensional features have a discriminative description to classify the liver diseases.
3.3 Comparison of Different Methods
We also make comparisons with current classification methods for liver ultrasound image [3, 6]. Sabih [3] chose liver lesion regions as ROI. This ROI was analyzed using Wavelet Packet Transform (WPT) and a number of statistical features were obtained. A multi-class linear SVM was then used for classification.
According to the advantages of existing ROI selection methods, Jeon [6] selected multiple ROI to represent well various ultrasonic appearances of liver lesions. In their view, multiple ROI could effectively hold a lot of used information and achieve good classification performance, regardless of features being used. We used five dimensional features from gray-level co-occurrence matrix, which achieved best classification results in paper [6].
As shown in Table 5, our method outperforms these two algorithms.
4 Conclusion
In this paper, we propose an innovate method to classify the focal liver lesions in ultrasound images. This method can automatically and accurately extract the lesion region to save time and labor. In feature extraction, we employ ultrasonic appearances of liver lesions. Specially, we design the mutation rate feature based on key point distribution. Combined with the GLCM and fractal features, we obtain 6 dimensional features which have a discriminative capability to distinguish focal liver lesions. Furthermore, we propose an improved SRC of DESR, which can expand and optimize the dictionary of sparse representation. Experimental results show that the proposed method can improve the classification performance significantly. The better accuracy and simple implementation make the proposed method beneficial to help physicians make a diagnosis, providing a realistic performance benchmark for further research efforts.
References
<http://www.cancer.gov/cancertopics/factsheet/Sites-Types/metastatic>
Santos, C.A., Brennan, D.C., Chapman, W.C., Fraser, V.J., Olsen, M.A.: Delayed-onset cytomegalovirus disease coded during hospital readmission in a multicenter, retrospective cohort of liver transplant recipients. Liver Transpl. 21, 581–590 (2015)
Sabih, D., Hussain, M.: Automated classification of liver disorders using ultrasound images. J. Med. Syst. 36, 3163–3172 (2012)
Owjimehr, M., Danyali, H., Helfroush, M.S.: Fully automatic segmentation and classification of liver ultrasound images using completed LBP texture features. In: Proceedings of Iranian Conference on Electrical Engineering (ICEE), pp. 1956–1960 (2014)
Alivar, A., Daniali, H., Helfroush, M.S.: Classification of liver diseases using ultrasound images based on feature combination. In: Proceedings of International Conference on Computer and Knowledge Engineering (ICCKE), pp. 669–672 (2014)
Jeon, J.H., Choi, J.Y., Lee, S., Ro, Y.M.: Multiple ROI selection based focal liver lesion classification in ultrasound images. Expert Syst. Appl. 40, 450–457 (2013)
Raghesh Krishnan, K., Radhakrishnan, S.: Focal and diffused liver disease classification from ultrasound images based on isocontour segmentation. IET Image Process. 9, 261–270 (2015)
Singh, M., Singh, S., Gupta, S.: An information fusion based method for liver classification using texture analysis of ultrasound images. Inf. Fusion 19, 91–96 (2014)
Wright, J., Yang, A.Y., Ganesh, A., Sastry, S.S., Ma, Y.: Robust face recognition via sparse representation. IEEE Trans. Pattern Anal. Mach. Intell. 31, 210–227 (2009)
Al Helal, A., Ahmed, K.I.: Rahman, M.S., Alam, S.K.: Breast cancer classification from ultrasonic images based on sparse representation by exploiting redundancy. In: Proceedings of International Conference on Computer and Information Technology (ICCIT), pp. 92–97 (2014)
Guo, Y., Wang, Y., Kong, D., Shu, X.: Automatic classification of intracardiac tumor and thrombi in echocardiography based on sparse representation. IEEE J. Biomed. Health Informat. 19, 601–611 (2015)
Aharon, M., Elad, M., Bruckstein, A.: K-SVD: an algorithm for designing over complete dictionaries for sparse representation. IEEE Trans. Sig. Process. 54, 4311–4322 (2006)
Wang, W., Li, J., Jiang, Y., Xing, Y., Xu, X.: An automatic energy-based region growing method for ultrasound image segmentation. In: Proceedings of International Conference on Image Processing (ICIP), pp. 1553–1557 (2015)
Mohanty, A.K., Beberta, S., Lenka, S.K.: Classifying benign and malignant mass using GLCM and GLRLM based texture features from mammogram. Int. J. Eng. Res. Appl. 1, 687–693 (2011)
Suganya, R., Rajaram, S.: Feature extraction and classification of ultrasound liver images using haralick texture-primitive features: application of SVM classifier. In: Proceedings of International Conference on Recent Trends in Information Technology (ICRTIT), pp. 596–602 (2013)
Kumar, S.S., Moni, R.S., Rajeesh, J.: Liver tumor diagnosis by gray level and contourlet coefficients texture analysis. In: Proceedings of International Conference on Computing, Electronics and Electrical Technologies (ICCEET), pp. 557–562 (2012)
Sarkar, N., Chaudhuri, B.B.: An efficient differential box-counting approach to compute fractal dimension of image. IEEE Trans. Syst. Man Cybern. 24, 115–120 (1994)
Allain, C., Cloitre, M.: Characterizing the lacunarity of random and deterministic fractal sets. Phys. Rev. A 44, 3552–3558 (1991)
Witkin, A.P.: Scale-space filtering: a new approach to multi-scale description. In: Proceedings of International Conference on Acoustics, Speech, and Signal Processing, pp. 150–153 (1984)
Alvarenga, A.V., Infantosi, A.F.C., Pereira, W.C.A., Azevedo, C.M.: Assessing the performance of morphological parameters in distinguishing breast tumors on ultrasound images. Med. Eng. Phys. 32, 49–56 (2010)
Chang, C.C., Lin, C.J.: LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 27 (2011)
Acknowledgements
This work was supported by the Natural Science Foundation of Guangdong Province #2015A030313212, Natural Science Foundation of China (NSFC) #61372007, #61171142, and the Science and Technology Planning project of Guangdong Province of China #2014B010111003, #2014B010111006, and the National Engineering Technology Research Center of Mobile Ultrasonic Detection #2013FU125X02. It was also supported in part by the National Natural Science Founding of China (U1636218), and Guangzhou Key Lab of Body Data Science (201605030011).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Wang, W., Jiang, Y., Shi, T., Liu, L., Huang, Q., Xu, X. (2017). Automatic Classification of Focal Liver Lesion in Ultrasound Images Based on Sparse Representation. In: Zhao, Y., Kong, X., Taubman, D. (eds) Image and Graphics. ICIG 2017. Lecture Notes in Computer Science(), vol 10667. Springer, Cham. https://doi.org/10.1007/978-3-319-71589-6_45
Download citation
DOI: https://doi.org/10.1007/978-3-319-71589-6_45
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-71588-9
Online ISBN: 978-3-319-71589-6
eBook Packages: Computer ScienceComputer Science (R0)