Abstract
Decreasing costs in genome sequencing have been paving the way for personalised medicine. An increasing number of individuals choose to undergo disease risk tests provided by medical units. However, it poses serious privacy threats on both the individuals’ genomic data and the tests’ specifics. Several solutions have been proposed to address the privacy issues, but they all suffer from high storage or communication overhead. In this paper, we put forward a general framework based on bloom filters, reducing the storage cost by 100x. To reduce communication overhead, we create index for encrypted genomic data. We speed up the searching of genomic data by 60x with bloom filter tree, compared to B\(_+\) tree index. Finally, we implement our scheme using the genomic data of a real person. The experimental results show the practicality of our scheme.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The first full sequencing of a human genome (“Human Genome Project”)Footnote 1 was completed in 2003, which takes 13 years, 3 billion dollars and involves more than 20 institutions world wide. Since then, numerous companies and research institutions have been moving toward more and more affordable and accurate technologies. For example, a genomics company called Illumina has brought the price of sequencing a human genome down to $1,000 in 2014, and the company claims that its newest machine can bring the cost down to $100 over the next few years. Advances in Whole Genome Sequencing (WGS) make genome testing available to the masses. At the same time, it stimulates the development of personalized healthcare such as predicting disease predisposition or response to the treatment. Some commercial companies (e.g., 23andMeFootnote 2) already provide low-cost risk tests to their customers for certain diseases.
However, genomic data disclosure brings great risk to personal privacy. Genomic information not only uniquely identifies its owner [1], but also reveals the genetic risk of diseases [2], leading to genetic discrimination (i.e., insurance or job hunting). Due to its hereditary nature, disclosure of an individual’s genome undermines the genomic privacy of his close relatives [3]. Although Genetic Information Non-discrimination Act (GINA) prohibits unauthorized use of genomic data, these kinds of laws are difficult to enforce. Furthermore, the genomes remain stable over time which cannot be revoked once made public [4]. Traditional anonymization methods have been proved to be ineffective for genomic data [5, 6]. As a result, current research focuses on privacy-enhancing technologies to protect genomic privacy. In general, we can classify the existing works into five main categories: (i) genome-wide association study (GWAS) [7], (ii) private string searching and comparisons [8], (iii) sequence alignments [9], (iv) private release of aggregate data [10], and (v) genomic tests [11].
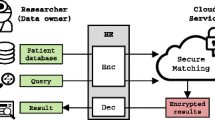
Disease Risk Computation Model. In this paper, we concentrate on protecting both the patients’ genomic data and tests’ specifics during disease risk tests. We adopt a similar model as [11] which involves a Patient (P), a Certified Institution (CI), a Storage and Processing Unit (SPU) and a Medical Unit (MU) as shown in Fig. 1. One patient provides his sample (e.g., his saliva) to the certified institution for sequencing. CI is a trusted party and responsible for sequencing and encrypting the patient’s genomic data. For efficiency and security, the encrypted genomic data is stored and processed at a centralized SPU. MU can be a hospital or a pharmaceutical company, which makes requests to SPU for computation on the genomic data. CI and P are honest parties, while MU and SPU are honest-but-curious (honestly follow the protocol, but try to infer private information).
Motivations. Single Nucleotide Polymorphism (SNP) is the most common DNA variation that occurs at a specific position in the genome.Footnote 3 SNP might have different states (i.e., homozygous or heterozygous). Position and state uniquely define a variation. There are approximately 50 million approved SNPs in the human population and each patient carries on average 4 million variants out of this 50 million. Different patients own different sets of 4 million SNPs. For a particular patient, all the 4 million SNPs are called real SNPs. The remaining SNPs (around 46 million) are considered as fictitious SNPs. Ayday et al. analyze the tradeoff between the amount of fictitious SNPs (storage cost) and the level of privacy [12]. The positions of SNPs are stored in plaintext at SPU, while the states are encrypted by homomorphic encryption.Footnote 4 Although the states are encrypted, SPU can still make inference about the states with the positions known, taking the correlation between SNPs into account (i.e., Linkage Disequilibrium [13] implies that SNPs at two positions are not independent of each other). As a result, they encrypt the positions in [11] as well. One drawback of [11] is that the squared value of each SNP must be stored at SPU to realize disease risk computation via homomorphic operations. Even worse, the protocol in [11] has two security weaknesses. The first one is that SPU can distinguish the requests for fictitious SNPs from the real ones, as they only store one ciphertext for all the fictitious SNPs. The second one is that it reveals how many SNPs are being tested. To remove the need of storing squares, Danezis and Cristofaro use an alternative encoding method [14]. Besides, their scheme encrypts all the possible SNPs of the human (50 million) for each patient. Consequently, SPU cannot differentiate requests for fictitious SNPs from those for real SNPs. Furthermore, they rely on padding, which means adding enough fictitious requests with zero weight to the test, to hide the number of SNPs relevant to the disease. Although they choose a small-ciphertext cryptosystem (AH-ECC [15]), downloading all the SNPs for a disease test is still unacceptable as it incurs high communication overhead. Besides, it depends heavily on the hardware security of a smartcard. In this paper, we aim to come up with a general framework to reduce the storage complexity of existing works, regardless of the underlying cryptosystem. For a disease risk test, we want MU to download only the SNPs related to the disease (padding included). Considering the large size of the dataset, it is impractical for SPU to search item by item. Therefore, a compact and efficient index structure is imperative.
Inspired by research on ciphertext retrieval [16], we treat the positions of SNPs as “keywords” and the corresponding states as “documents”. Bloom filter [17] is a space-efficient probabilistic data structure to test whether an element is a member of a set. In our scheme, we let CI encrypt the positions of SNPs and insert them into bloom filters. For 50 million possible SNPs, we create 0.5 million bloom filters and each bloom filter contains 100 SNPs with the same state.Footnote 5 To avoid searching all the bloom filters sequentially, we organize them with a tree structure. Then CI uploads only the bloom filter tree to SPU. As SPU does not store the encrypted positions, bloom filters take up little space. For example, the size of a bloom filter which contains 1 million elements is around 1.7 megabytes (with false positive rate \(0.1\%\)). We link one ciphertext to each bloom filter at the leaf node level to represent the encrypted states of SNPs. In [11, 14], they store one ciphertext (encrypted state) for each position of SNPs. However, we store one ciphertext for 100 positions of SNPs. Without considering the small-size index structure, we reduce the storage overhead by 100x. On the other hand, bloom filter provides fast searching speed. For example, we only need to compute ten hash functions to check whether an element belongs to a bloom filter which contains 1 million elements (with false positive rate \(0.1\%\)). If MU wants to conduct disease risk test for a patient, MU will send a list of encrypted positions to SPU. Then SPU checks the bloom filter tree hierarchically and returns the corresponding encrypted states to MU if contained. The disease risk is computed on the MU’s side via homomorphic operations. Once the computation is done, the final disease risk is disclosed to MU under the authorization of the patient. The main contributions of our work are summarized as follows.
-
(1)
We put forward a general framework to reduce the storage overhead of existing works by 100x, regardless of the underlying cryptosystem. To be specific, we insert 100 positions into a bloom filter and link one ciphertext to the bloom filter to represent the encrypted states of SNPs.
-
(2)
For genetic disease risk test, we are the first to create index for the encrypted SNPs stored at SPU. We organize bloom filters with a tree structure. Compared to B\(_+\) tree index, we speed up the searching of SNPs by 60x.
-
(3)
We implement our scheme with SNPs of a real person. The experimental results show the practicality of our scheme.
2 Related Work
Ziegeldorf et al. perform string matching with bloom filters [18] on the Track 3 dataset provided by the iDASH Secure Genome Analysis Competition. They assume n patients have SNPs at the same m locations, without considering the fact that different patients own different sets of 4 million SNPs. Our paper handles with the positions and states of the SNPs separately. We focus on the storage and searching of SNPs [11, 12, 14] in this paper. But beyond that, there exists some related work worthy to be discussed. Since the genome contains the most sensitive information, how and where should a patient’s genomic data be stored have always been controversial. In the cloud? In a physician’s office? On a smartphone? All have strengths and limitations in terms of portability, capacity, computing power and privacy. One personal genomic toolkit called GenoDroid is implemented on the Android platform [19]. To reduce the computation on mobile phones, the genomic data and certain cryptographic operations should be preprocessed on a laptop. On the other hand, although the SNPs are encrypted at SPU, a honest-but-curious SPU learns which SNPs are accessed, how often SNPs are used. Therefore, SPU might try to infer the nature of the ongoing test. To hide the access pattern, [20] uses Oblivious RAM (ORAM) to store DNA in several small encrypted blocks. The blocks are accessed in an oblivious manner. However, the usage of ORAM will inevitably incur large cost due to periodically reshuffle process, which is impractical to implement in real-life scenarios. Furthermore, as MU knows the SNP weights, the risk computation can be seen as a linear equation where the states of SNPs are the unknowns. MU can launch brute-force attack. For example, we assume 10 SNPs are used in the disease risk test. Each SNP only has 3 different states. Therefore, MU can try all the \(3^{10}\) combinations of SNPs. If one of these potential end-results matches the actual end-result of the test, the MU actually learns the states of the relevant SNPs. One simple obfuscation method is to provide the end-result of the genetic test to MU as a range. Therefore, Ayday et al. discussed how to use privacy-preserving integer comparison to report the range of genetic test [21]. Provided that the result range is divided into 4 smaller ranges [0, 0.25), [0.25, 0.50), [0.50, 0.75) and [0.75, 1], SPU compares the encrypted disease risk with those pre-defined boundaries to determine the range that the test result falls in. Moreover, they take into account clinical and environmental data to compute the final disease risk, such as hypertension, age, smoking and family disease history. The above techniques can be combined with our scheme proposed in this paper.
3 Preliminaries
3.1 Genomics Background
The human genome is encoded in double stranded DNA molecules consisting of two complementary polymer chains. Each chain consists of simple units called nucleotide or “bases”, adenine (A), cytosine (C), guanine (G), thymine (T). Specifically, A/T and C/G are complementary pairs, respectively. The human genome consists of approximately three billion nucleotides, of which only around \(0.1\%\) of one individual’s DNA is different from others due to genetic variation. The remaining \(99.9\%\) of the genome is identical between any two individuals.
What is SNP? Single Nucleotide Polymorphism (SNP) is the most common DNA variation. SNP is a variation in a single nucleotide that occurs at a specific position in the genome. For example, at a specific position of two DNA fragments in Fig. 2, the nucleotide C may appear in most individuals, but in a minority of individuals, the position is occupied by base T. In this case, we say there is a SNP at this position and we call the two possible nucleotide variations (C or T) alleles. Each individual carries two alleles at each SNP (one inherited from the father and one from the mother). There are approximately 50 million approved SNPs in the human population and each patient carries on average 4 million variants. Different patients own different sets of 4 million SNPs. For a particular patient, all the 4 million SNPs are called real SNPs and the remaining SNPs are considered as fictitious SNPs.
Relationship Between SNPs and Genetic Diseases: It is shown by Multiple Genome-Wide Association Studies (GWAS) that a patient’s susceptibility to genetic diseases can be predicted from SNPs. Assume that the SNP in Fig. 2 (with alleles C and T) is relevant to a disease and T is the one carrying the disease risk, then an individual with TT has the highest risk to develop the disease. CT implies lower disease risk. And individuals with CC are least likely to suffer from the disease. We use \(\{0, 1, 2\}\) to represent the number of risk alleles. Furthermore, we adopt the encoding scheme in [14] to express \(\{0, 1, 2\}\) as \(\{100, 010, 001\}\), as binary encoding of SNPs is able to remove non-linear operations (i.e., squaring).
Disease Risk Computation: There are different functions for computing disease risk. One popular method is weighted averaging, which computes the predicted susceptibility by weighting the SNPs by their contributions. For each SNP\(_i\), the contribution amount \(w_i\) is known by the medical unit, determined by previous studies on case and control groups. To be specific, \(w_i\) relies on position contribution \(p_i\) and state contribution \(c_i\), and we have \(w_i=p_ic_i\). Assume that we denote the set of SNPs of a user associated with disease X as I, the risk can be calculated by \(S=\frac{\sum w_iI_i}{\sum p_i}\).
3.2 Bloom Filters
Bloom filter [17] is a space-efficient probabilistic data structure to test whether an element is a member of a set. It is represented by a m-bit array \(B\in \{0,1\}^m\). The bits of an empty bloom filter are all set to 0. Before inserting elements, we choose k independent hash functions \(h_1,h_2,\cdots ,h_k\) which map the elements into the range [1, m]. To add an element \(e_i\) to the bloom filter, we calculate k hash values \(h_1(e_i),h_2(e_i),\cdots ,h_k(e_i)\) and set the bits of B corresponding to those hash values to 1. Similarly, to test whether \(e_i\in B\), we check the bits at positions \(h_1(e_i),h_2(e_i),\cdots ,h_k(e_i)\). If all the bits are 1, then \(e_i\) is considered as a member of the set. Otherwise, \(e_i\) is not contained in the set. In Fig. 3, we provide an example. Firstly, \(x_1\) and \(x_2\) are added to one bloom filter using three hash functions. Then we check whether \(y_1\) or \(y_2\) is in the set, and we can conclude that the set contains \(y_2\) instead of \(y_1\). Due to hash collisions, bloom filter may produce false positive, where it suggests that an element is in the set even though it is not. The false positive probability p is determined by the number of hash functions k, the number of added elements n and the length m of the bloom filter.
Given a fixed n, if we want to keep p below a threshold \(\epsilon \), we need to set m to be \(m \ge n\ log_2\ e \cdot log_2\ ({\frac{1}{\epsilon }})\) where e is the base of the natural logarithm and \(log_2\ e \approx 1.44\) [22]. The optimal value for k is \(\frac{mln(2)}{n}\). For many applications, false positives may be acceptable as long as their probability is sufficiently small.
3.3 Homomorphic Encryption
Homomorphic encryption performs computation on ciphertexts, and generates an encrypted result which matches the result of operations performed on the plaintexts when decrypted. Homomorphic encryption can be classified to three categories: (i) Fully Homomorphic Encryption (FHE), (ii) Somewhat homomorphic encryption (SWHE), and (iii) Partially Homomorphic Encryption (PHE). We focus on PHE in this paper and show an additively homomorphic encryption scheme - the modified paillier [23] in Fig. 4. The additive homomorphism is denoted as \(E(m_1+m_2)=E(m_1)E(m_2)\)Footnote 6.
4 Our Scheme
In this paper, we aim to put forward a general framework to reduce the storage overhead, regardless of the underlying cryptosystem. For a disease risk test, we want MU to download only the SNPs related to the disease (padding included). Thus, we are supposed to build index to make SPU efficiently select SNPs. Recall that position and state uniquely define a SNP, we should select cryptosystems for them separately. In Sect. 4.1, we provide you with the building blocks required to construct a privacy-preserving disease risk test protocol. Then we demonstrate our scheme in Sect. 4.2.
4.1 Building Blocks
Encryption of SNPs’ Positions. As position information of SNPs is also sensitive, CI needs to decide on a cryptosystem to encrypt the positions. We provide two options which apply to different scenarios - a symmetric encryption scheme (i.e., AES), and a public key encryption scheme (i.e., ECC). Symmetric scheme needs the help of P to submit MU’s requests to SPU. Whereas, public key encryption scheme enables MU to submit requests independently.
Encryption of SNPs’ States. Any additively homomorphic encryption scheme can be used to encrypt the states of SNPs. Although [14] shows that AH-ECC cryptosystem provides smaller ciphertexts and faster operations, decryption requires to compute discrete log. As the discrete log problem is assumed to be hard, we have to maintain a pre-computed table of discrete logarithms (which is only practical for small ranges of plaintext). Therefore, in this paper, we choose modified paillier cryptosystem (Fig. 4) for ease of implementation.
Storage Overview. For 50 million possible SNPs, we create 0.5 million bloom filters. Each bloom filter contains 100 SNPs with the same state. In order to represent the encrypted states of SNPs, we link one ciphertext to each bloom filter. We show the storage architecture in Fig. 5. On the left side, we store the encrypted states of the SNPs. On the right side, we store the bloom filters. There is a one-to-one correspondence between the encrypted state and the bloom filter at each row. To avoid searching all the bloom filters sequentially, we organize them with a tree structure. To make it clear, we show a binary tree example in Fig. 6. At the leaf node level, we store the 0.5 million bloom filters and the corresponding ciphertexts. At level \(i-1\), we store bloom filters which double the size of the bloom filters at level i. However, we want to point out that our scheme is not restricted to binary tree. Any multi-branch tree can be used. For a k-branch tree, the size of bloom filters at level \(i-1\) is k times the size of bloom filters at level i.
Why a Bloom Filter Contains 100 Positions? The decrease of storage overhead depends on the capacity of each bloom filter. With the capacity increases, the storage overhead decreases. As there are three possible states for each SNP, we only need three bloom filters in the extreme case, with each bloom filter at maximum capacity. In this case, we get the maximum storage overhead decrease. However, the length of bloom filter reveals the frequency of different states of SNPs. Therefore, we need to fix a constant capacity for the bloom filters. On the other hand, there is one security weakness that SPU will know requests made to the same bloom filter might have the same state. Supposed that there are b bloom filters, and MU requests for t real SNPs, the probability that any two requests appear on the same bloom filter is \(C_t^{2}/b\). For fixed t, the above probability decreases with b increases. The number of real SNPs for a particular disease is quite small (i.e., less than 60)Footnote 7. For example, calculating the susceptibility of Alzheimer’s disease requires only 10 SNPs [24] and coronary artery disease risk computation includes 23 SNPs [25]. With 0.5 million bloom filters, the probability that any two different requests appear on the same bloom filter is at most \(C_{60}^{2}/(5\times 10^5)=0.35\%\). Therefore, we choose the constant capacity 100 for the bloom filters to achieve a balance between the decrease of storage overhead and privacy.
4.2 Our Construction
We present the steps of a privacy-preserving disease risk test in this section. We assume that the cryptographic keys are generated by a trusted authority and distributed to the involved parties at the initialization period.
-
Step 1: P provides his sample to CI for sequencing.
-
Step 2: Firstly, CI randomizes the positions of SNPs. Then CI encrypts the positions by symmetric encryption (or public key encryption) and inserts them into the bloom filters. Moreover, CI encrypts the states of SNPs by modified paillier cryptosystem and links one ciphertext to each bloom filter. Then CI organizes the bloom filters with a tree structure. CI uploads the bloom filter tree and the corresponding ciphertexts to SPU.
-
Step 3: P goes to the MU to take disease risk test. MU pads some fictitious positions among real ones to hide the number of SNPs involved in the test.
-
If symmetric encryption is adopted in Step 2, MU has to send the positions (padding included) to the patient first. Then patient encrypts the positions and sends the encrypted positions to SPU.
-
If public key encryption is used in Step 2, it is possible for MU to make requests to SPU directly without the help of P. To prevent SPU from checking whether an element is contained in a bloom filter without P’s authorization, we make use of keyed hashing when creating bloom filters. If P agrees to conduct the disease risk test, then MU gets the hash keys.
-
-
Step 4: SPU receives the encrypted positions from P (or MU). SPU will check the index tree of bloom filters hierarchically. If the encrypted position lies in a bloom filter, then SPU will return the ciphertext linked to this bloom filter to MU. If two or more requests occur in the same bloom filter, SPU should re-encrypt the ciphertext before returning to MU (Eq. (4)). Furthermore, we require SPU to return the encrypted states according to the order of MU’s requests.
-
Step 5: Once getting the encrypted states of requested SNPs, MU computes the disease risk via homomorphic operations based on weighted averaging.
-
Step 6: The encrypted final result is sent to P. After decryption, the result is return back to MU.
Extensions: Disease risk computation is based on weighted averaging. Apart from genomic data, clinical and environmental data can be integrated into our scheme. For better security, we can output the disease risk as a range. Ayday et al. demonstrate how to use privacy-preserving integer comparison [26] to report the range of genetic test [21], which can be easily incorporated into our scheme. Besides, if the secret key of a patient is divided into two secret shares and distributed to SPU and MU, the final result can be revealed to MU after two rounds of partial decryption without the patient’s help.
5 Privacy Analysis
The SNPs of a patient contain sensitive information. Homomorphic encryption makes it possible to compute disease risk for the patients without revealing the true values of SNPs. At the same time, the medical unit (i.e., pharmaceutical company) might consider the test specifics as trade secret. Hence, MU does not want to make public the weights (or even the number) of the disease test. Therefore, we conduct privacy analysis of our proposed scheme in term of these two aspects. The MU will download the requested encrypted SNPs and compute the disease risk locally. Therefore, the privacy of test weights preserves. Recall that CI and P are honest parties, SPU and MU are honest-but-curious (Fig. 1), we consider two kinds of attacks: (i) an attacker at MU trying to know the SNPs of the patient (type-1 attack). (ii) an attacker at SPU inferring the requests from MU and the SNPs of the patient (type-2 attack).
Security Under Type-1 Attack: The MU makes requests to SPU for SNPs of a patient to conduct disease risk test. SPU returns the corresponding encrypted states of the requested SNPs to MU. Even different requests occur in the same bloom filter, SPU will re-encrypt the ciphertext. Therefore, the ciphertexts are indistinguishable among different requests from the point of view of MU. Security under type-1 attack relies on the security of the underlying cryptosystem that encrypts the states. In our scheme, any additively homomorphic cryptosystem can be used. As long as the underlying cryptosystem is semantic secure, it is unable for the attacker at MU to infer any additional information about the plaintext with the ciphertext known. Most of the additively homomorphic cryptosystems base their security on assumptions relying on deciding residuosity. But the two cryptosystems we mentioned in Sect. 4.1 base on non-residuosity-related decisional assumption. To be specific, the security of modified paillier cryptosystem [23] is based on decisional Diffie-Hellman assumption. The security of AH-ECC is based on the intractability of Elliptic Curve Discrete Logarithm Problem (ECDLP). Due to space limit, we do not go into details here.
Security Under Type-2 Attack: The MU requests for SNPs with encrypted positions. As the positions are encrypted, it is impossible for SPU to know the true positions as long as the underlying cryptosystem is secure. Taking AES for an example, the key is kept secret by the patient. Anyone without the secret key cannot deduce anything from the ciphertexts. Moreover, MU pads some fictitious positions among the real positions. In this way, it successfully hide the number of the requested SNPs, which prevents the SPU from speculating the disease type according to the number of involved SNPs. We then show that the requests for SNPs are distributed uniformly among different bloom filters. First, the positions are randomized at CI before being inserted into the bloom filters. Second, we choose hash functions which map the inputs as evenly as possible over the output range to create bloom filters. In other words, every hash value in the output range is generated with roughly the same probability. Therefore, the distribution of ones in different bloom filters reveals nothing about its inputs. Recall that we link one ciphertext to each bloom filter, there is one security weakness that SPU will know requests made to the same bloom filter might have the same state. In this paper, we create 0.5 million bloom filters for 50 million possible SNPs to achieve a trade-off between the decrease of storage overhead and privacy. If co-occurrence does happen, we want to point out that even though SPU might know requests on the same bloom filter are likely to have the same state, it cannot know whether it is a real or fictitious request or what the state is.
6 Experimental Evaluation
To evaluate the proposed scheme, we implemented it on our PC which is Windows 7 Enterprise 64-bit Operating System with Intel(R) Core(TM) i5 CPU (4 cores), 3.4 GHz and 16 GB memory. To provide platform-independence, we use the Java programming language.
We downloaded the SNPs of a real individual from openSNP.Footnote 8 There are 985,949 (around 1 million) SNPs in our dataset. We run experiments on this smaller dataset (compared to 50 million) to measure the searching speed and storage cost. We use the CCM mode of AES to encrypt the positions of SNPs, and modified paillier cryptosystem to encrypt the states of SNPs with 1024-bit security parameter. In order to facilitate the subsequent processing, we saved the encrypted positions and states into a MySQL 5.5 database. To uniquely locate a SNP, we combine rsid (reference SNP cluster id), chromosome number and offset to denote the position of each SNP. There are around 1 million SNPs in our dataset. It is notoriously slow to search for one particular encrypted position without index. MySQL does provide us with a way to create index for the tables based on B\(_+\) three, which performs well in running range search (i.e., [1, 100]). However, the encrypted positions of SNPs related to a particular disease may be scattered throughout the space. In this scenario, bloom filter is better in terms of searching speed and storage. Recall that we store 100 SNPs in each bloom filter, we have around \(10^4\) bloom filters for our dataset. To facilitate presentation, we organize the bloom filters with 10-branch tree. The number of bloom filters at each level and the capacity of each bloom filter are shown in Table 1. We show the time to search requested SNPs at SPU based on B\(_+\) tree or bloom filter tree in Fig. 7, varying the number of SNPs from 1 to 1000. For B\(_+\) tree index, the searching time increases from 130 ms to 3200 ms with increasing number of SNPs. By contrast, for bloom filter tree index, the searching time increases from 6.4 ms to 52 ms approximately linearly. On average, we speed up the searching by 60x.
Since bloom filter may cause false positive, we also measure the effect of false positive rate on the size of our index (bloom filter tree). Given a fixed n, if we want to keep p below a threshold \(\epsilon \), we need to set m to be \(m \ge n\ log_2\ e \cdot log_2\ ({\frac{1}{\epsilon }})\) (see Sect. 3.2). For 1 million elements, the size of our index under false positive rate \(1\%\), \(0.1\%\) and \(0.01\%\) is 4.84 MB, 6.91 MB and 9.68 MB, respectively. The number of hash functions used in each bloom filter is 7, 10 and 14, respectively. In comparison, the size of B\(_+\) three index generated by MySQL is 48.69 MB. Therefore, our bloom filter tree index not only speeds up the searching of SNPs but also is space-efficient. All the existing works store one encrypted state for each SNP (i.e., [11, 14]). To be specific, one encrypted state corresponds to one position. But in this paper, we insert 100 positions with the same state into one bloom filter and link one ciphertext to the bloom filter. Therefore, one encrypted state corresponds to 100 SNP positions. This is the reason why we can reduce the storage overhead by 100x. It is shown in [14] that the encryption of 50 million SNPs takes about 100 GB by modified paillier cryptosystem, and 4.5 GB by AH-ECC cryptosystem (under 112-bit security parameter). As a result, our scheme makes a big difference in storage overhead regardless of the underlying cryptosystem.
7 Discussion and Conclusions
In this paper, we assume that P and CI are trusted, SPU and MU are honest-but-curious. If we allow MU to be dishonest (arbitrarily deviates from the protocol), it is likely that MU will launch illegal requests (i.e., with one weight of value 1, the remaining is 0) to SPU to infer the patient’s SNPs. We can make a compromise between the privacy of MU (i.e., tests’ specifics) and the privacy of patient’s SNPs [27]. MU is forced to reveal iteratively some weights to the SPU, until the SPU is convinced that the ongoing test is legitimate.
To conclude, we build an efficient index tree for the encrypted SNPs with bloom filters. We speed up the searching of SNPs by 60x compared to B\(_+\) tree index. Moreover, we successfully reduce the storage cost by 100x regardless of the underlying cryptosystem. We implement our scheme on SNPs of a real patient, the experimental results highlight the practicality of our scheme. Due to the space-efficient property of bloom filter, our future work is to optimize our scheme and deploy it on smartphones without a centralized SPU.
Notes
- 1.
The human genome project, https://www.genome.gov/12011238/an-overview-of-the-human-genomeproject/.
- 2.
https://www.23andme.com. Accessed on 8/Sep/2017.
- 3.
Refer to the explanation of SNP in Sect. 3.1.
- 4.
See the definition of homomorphic encryption in Sect. 3.3.
- 5.
We discuss the reason for choosing 100 in Sect. 4.1.
- 6.
\(m_1, m_2\) are plaintexts and \(E(m_1)\), \(E(m_2)\) are their corresponding ciphertexts.
- 7.
http://www.eupedia.com/genetics/cancer_related_snp.shtml, accessed on 9 Sep 2017.
- 8.
https://opensnp.org, accessed on 16 Aug 2017.
References
Homer, N., Szelinger, S., Redman, M., Duggan, D., Tembe, W., Muehling, J., Pearson, J.V., Stephan, D.A., Nelson, S.F., Craig, D.W.: Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 4(8), e1000167 (2008)
Altshuler, D., Daly, M.J., Lander, E.S.: Genetic mapping in human disease. Science 322(5903), 881–888 (2008)
Humbert, M., Ayday, E., Hubaux, J.-P., Telenti, A.: Addressing the concerns of the lacks family: quantification of kin genomic privacy. In: Proceedings of the 2013 ACM SIGSAC Conference on Computer and Communications Security, pp. 1141–1152. ACM (2013)
Erlich, Y., Narayanan, A.: Routes for breaching and protecting genetic privacy. Nat. Rev. Genet. 15(6), 409–421 (2014)
Malin, B.A.: An evaluation of the current state of genomic data privacy protection technology and a roadmap for the future. J. Am. Med. Inform. Assoc. 12(1), 28–34 (2005)
Shringarpure, S.S., Bustamante, C.D.: Privacy risks from genomic data-sharing beacons. Am. J. Hum. Genet. 97(5), 631–646 (2015)
Zhang, Y., Blanton, M., Almashaqbeh, G.: Secure distributed genome analysis for gwas and sequence comparison computation. BMC Med. Inform. Decis. Mak. 15(5), S4 (2015)
Perl, H., Mohammed, Y., Brenner, M., Smith, M.: Privacy/performance trade-off in private search on bio-medical data. Future Gener. Comput. Syst. 36, 441–452 (2014)
Chen, Y., Peng, B., Wang, X.F., Tang, H.: Large-scale privacy-preserving mapping of human genomic sequences on hybrid clouds. In: NDSS (2012)
Zhou, X., Peng, B., Li, Y.F., Chen, Y., Tang, H., Wang, X.F.: To release or not to release: evaluating information leaks in aggregate human-genome data. In: Atluri, V., Diaz, C. (eds.) ESORICS 2011. LNCS, vol. 6879, pp. 607–627. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-23822-2_33
Ayday, E., Raisaro, J.L., Hubaux, J.-P., Rougemont, J.: Protecting and evaluating genomic privacy in medical tests and personalized medicine. In: Proceedings of the 12th ACM Workshop on Workshop on Privacy in the Electronic Society, pp. 95–106. ACM (2013)
Ayday, E., Raisaro, J.L., Hubaux, J.-P.: Personal use of the genomic data: privacy vs. storage cost. In: 2013 IEEE Global Communications Conference (GLOBECOM), pp. 2723–2729. IEEE (2013)
Falconer, D.S., Mackay, T.F.C., Frankham, R.: Introduction to Quantitative Genetics. Trends in Genetics, vol. 12, no. 7, 4th edn, 280 p. Elsevier Science Publishers (Biomedical Division), Amsterdam (1996)
Danezis, G., De Cristofaro, E.: Fast and private genomic testing for disease susceptibility. In: Proceedings of the 13th Workshop on Privacy in the Electronic Society, pp. 31–34. ACM (2014)
Ugus, O., Westhoff, D., Laue, R., Shoufan, A., Huss, S.A.: Optimized implementation of elliptic curve based additive homomorphic encryption for wireless sensor networks. arXiv preprint arXiv:0903.3900 (2009)
Huang, R.W., Gui, X.L., Yu, S., Zhuang, W.: Research on privacy-preserving cloud storage framework supporting ciphertext retrieval. In: 2011 International Conference on Network Computing and Information Security (NCIS), vol. 1, pp. 93–97. IEEE (2011)
Bloom, B.H.: Space/time trade-offs in hash coding with allowable errors. Commun. ACM 13(7), 422–426 (1970)
Ziegeldorf, J.H., Pennekamp, J., Hellmanns, D., Schwinger, F., Kunze, I., Henze, M., Hiller, J., Matzutt, R., Wehrle, K.: BLOOM: bloom filter based oblivious outsourced matchings. BMC Med. Genomics 10(2), 44 (2017)
De Cristofaro, E., Faber, S., Gasti, P., Tsudik, G.: Genodroid: are privacy-preserving genomic tests ready for prime time? In: Proceedings of the 2012 ACM Workshop on Privacy in the Electronic Society, pp. 97–108. ACM (2012)
Karvelas, N., Peter, A., Katzenbeisser, S., Tews, E., Hamacher, K.: Privacy-preserving whole genome sequence processing through proxy-aided ORAM. In: Proceedings of the 13th Workshop on Privacy in the Electronic Society, pp. 1–10. ACM (2014)
Ayday, E., Raisaro, J.L., Laren, M., Jack, P., Fellay, J., Hubaux, J.-P.: Privacy-preserving computation of disease risk by using genomic, clinical, and environmental data. In: Proceedings of USENIX Security Workshop on Health Information Technologies (HealthTech 2013), no. EPFL-CONF-187118 (2013)
Broder, A., Mitzenmacher, M.: Network applications of bloom filters: a survey. Internet Math. 1(4), 485–509 (2004)
Bresson, E., Catalano, D., Pointcheval, D.: A Simple public-key cryptosystem with a double trapdoor decryption mechanism and its applications. In: Laih, C.-S. (ed.) ASIACRYPT 2003. LNCS, vol. 2894, pp. 37–54. Springer, Heidelberg (2003). https://doi.org/10.1007/978-3-540-40061-5_3
Seshadri, S., Fitzpatrick, A.L., Arfan Ikram, M., DeStefano, A.L., Gudnason, V., Boada, M., Bis, J.C., Smith, A.V., Carrasquillo, M.M., Lambert, J.C., et al.: Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303(18), 1832–1840 (2010)
Rotger, M., Glass, T.R., Junier, T., Lundgren, J., Neaton, J.D., Poloni, E.S., Van’t Wout, A.B., Lubomirov, R., Colombo, S., Martinez, R., et al.: Contribution of genetic background, traditional risk factors, and HIV-related factors to coronary artery disease events in HIV-positive persons. Clin. Infect. Dis. 57(1), 112–121 (2013)
Erkin, Z., Franz, M., Guajardo, J., Katzenbeisser, S., Lagendijk, I., Toft, T.: Privacy-preserving face recognition. In: Goldberg, I., Atallah, M.J. (eds.) PETS 2009. LNCS, vol. 5672, pp. 235–253. Springer, Heidelberg (2009). https://doi.org/10.1007/978-3-642-03168-7_14
Barman, L., Graini, E., Raisaro, J.L., Ayday, E., Hubaux, J.-P., et al.: Privacy threats and practical solutions for genetic risk tests. In: 2nd International Workshop on Genome Privacy and Security (GenoPri 2015), no. EPFL-CONF-207435 (2015)
Acknowledgement
This project is partially supported by a collaborative research grant (RGC Project No. CityU C1008-16G) of the Hong Kong Government.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Zhang, J., Zhang, L., He, M., Yiu, SM. (2018). Privacy-Preserving Disease Risk Test Based on Bloom Filters. In: Qing, S., Mitchell, C., Chen, L., Liu, D. (eds) Information and Communications Security. ICICS 2017. Lecture Notes in Computer Science(), vol 10631. Springer, Cham. https://doi.org/10.1007/978-3-319-89500-0_41
Download citation
DOI: https://doi.org/10.1007/978-3-319-89500-0_41
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-89499-7
Online ISBN: 978-3-319-89500-0
eBook Packages: Computer ScienceComputer Science (R0)