Abstract
Janus nanoparticles have surfaces with two or more distinct physical properties, allowing different types of chemical properties to occur on the same particle and thus making possible many unique applications. It is necessary to investigate the interaction between proteins and Janus nanoparticles (NPs), which are two typical building blocks for making bio-nano-objects. Here we computed the phase diagrams for an \(\alpha \)+\(\beta \) protein(GB1) and Janus NP using coarse-grained model and molecular dynamics simulations, and studied how the secondary structures of proteins, the binding interface and kinetics are affected by the nearby NP. Two phases were identified for the system. In the folded phase, the formation of \(\beta \)-sheets are always enhanced by the presence of NPs, while the formation of \(\alpha \)-helices are not sensitive to NPs. The underlying mechanism of the phenomenon was attributed to the geometry and flexibility of the \(\beta \)-sheets. The knowledge gained in this study is useful for understanding the interactions between proteins and Janus NP which may facilitate designing new bio-nanomaterials or devices.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Nanomaterials have attracted attentions from different branches of science and technology, such as physics, chemistry, biology and medicine. Nanomaterials are different from their bulk counterparts in that they have tiny size and high volume to surface ratio, which lead to special chemical, electrical, optical properties, high catalyst efficiencies, and some other intriguing features [1,2,3]. Of the same importance and interest are the bio-materials formed of such as polymers, peptides, proteins, or nucleic acids. Biomaterials are easy to be designed at molecular level and able to self- or co-assemble into highly organized structures [2]. Nowadays, it has become a thriving research area to combine nanomaterials and biomaterials and make novel functional materials or tiny devices for drug delivery, bioimaging, sensing, diagnosing, or more speculatively, nanomachines and nanorobots [4,5,6]. Among all of the possible building blocks for such purposes, proteins represent excellent ones for they have sophisticated structures at nanoscale dimensions, rich chemistry and versatile enzymatic activities. Therefore, it is necessary to study how proteins interact with nanomaterials such as small nanoparticles (NPs).
Early studies of NPs are mainly concerned with the effects of the physical properties of NPs, such as size and shape, on the structure of protein [7,8,9]. For example, Shang et al. [8] studied the changes in the thermodynamic stability of the ribonuclease A when it adsorbed on the silicon NPs. The results show that the larger the size of the NPs, the greater the effect on the thermodynamic stability of the protein. Gagner et al. [9], studied the influence of protein structure by the shape of NPs when the protein adsorbed on gold NPs. The NPs used in the experiment were spherical gold NPs (diameter 10.6 ± 1 nm) and columnar gold NPs (bottom diameter 10.3 ± 2 nm), respectively, and the proteins were lysozyme and chymotrypsin. The results show that the concentration of protein molecules adsorbed on the surface of the columnar gold NPs is higher for the allogeneic protein. After that, the researchers found that the surface chemical properties of the NPs could produce a more abundant effect. For example, Rocha et al. [10] studied the effects of fluorinated NPs and hydrogenated NPs on the aggregation of \(A\beta \) protein The results show that fluorinated NPs could induce the transformation of \(\beta \)-sheets to \(\alpha \)-helix, thus inhibiting aggregation, while hydrogenated NPs could induce random curling to form \(\beta \)-sheets, thereby promoting the occurrence of aggregation.
With the development of research, it has been found that one NP with different surface characteristics can play a variety of functions and have wider application value [11,12,13,14,15]. Roh et al. [16] designed a Janus NP, which was synthesized by two different materials. Half of the material emitted green fluorescence after adding fluorescein isothiocyanate labeled dextran, and the other half of the material emitted red fluorescence after adding rhodamine B labeled dextran. The experimental results show that after the mixture of two different modal molecules and Janus NPs, half of the NPs emitted green fluorescence and half of them emitted red fluorescence, that is, the two molecules could combine with the surface of particles with different properties respectively. This indicates that Janus NPs can indeed carry different molecules, which can be applied to molecular detection or drug transport. Further experiments show that the surface of Janus NPs can be selectively modified by using the characteristics of the interaction between Janus NPs and different molecules, so as to design more kinds of NPs. Honegger et al. [17] synthesized gold NPs with polystyrene and silica respectively, and mixed the Janus NPs with proteins. The results show that different proteins can accurately adsorb on the surface of a specific material. This further illustrates the feasibility of the application of Janus NPs in the field of molecular detection.
Although a lot of work has been done to explore the synthesis and application of Janus NPs, the understanding on the effect of Janus NPs on the surrounding protein is still limited. There is still no systematic investigation on how they interact with each other exactly. Many interactions, such as the hydrophobic force, Coulomb force, hydrogen-bond, polarizability and lone-pair electrons, may contribute to the final result [18]. While in most experiments, which interaction is the most relevant factor governing a specific observation is usually not clear. Thus, a close collaboration between experimentalists and theorists is preferred [19,20,21]. In this work we study the adsorption process of a \(\alpha \)+\(\beta \) protein on a spatially nearby Janus NP based on a coarse-grained model and molecular simulations. The coarse-grained model allows us to explore a large parameter space spanned by different surface chemical properties and NP sizes and different strengths of protein-NP interactions. In this way we are able to systematically examine the effect of NPs on proteins and seek whether there are any general principles. The conformational phase diagrams for protein-NP combination, the binding kinetics and the changes of protein structures on adsorption are investigated. The results are compared with experiments and the underlying mechanisms are then discussed.
2 Models and Methods
2.1 Models of the Proteins and Janus NP
An \(\alpha \)+\(\beta \) protein GB1 (PDB ID: 3GB1) was studied to investigate how a protein is affected by the presence of a Janus NP in its proximity. The sequence length of the protein is 56, and the radius of gyration of its native tertiary structures is 1.04. The Janus NP that interacts with the proteins was treated as a rigid spherical bead with half surface hydrophobic and half surface hydrophilic.
To characterize the protein adsorption on NPs, we first define the contact between protein and NPs. A contact between residue i and NPs is deemed to be formed if the distance between the residue and the NP surface is less than 1.2\(d_0\), where coefficient 1.2 is used to reflect the thermal fluctuations, following the strategy in the literature [22]. We also define the fraction of NP surface covered by protein as \(S=1-SASA/S_{NP}\), where \(S_{NP}\) is the surface area of the NP and SASA is its solvent accessible surface area that is not covered by protein, obtained by rolling a small probing bead with a radius of  on the NP surface.
on the NP surface.
The status of the interacting protein and the NP is denoted by a two-letter word, for example, AF, DF, AU and WU. The first letter describes the binding status of the protein on NP, which can be desorbed (D), adsorbed (A) or wrapped (W), corresponding to the parameter range \(S<0.1\), \(0.1<S<0.8\), or \(S>0.8\), respectively. The second letter indicates the folding status of the protein C either the folded state (denoted as F, with \(Q>0.75\)) or the unfolded state (denoted as U, with \(Q<0.75\)). The value of 0.75 is deliberately chosen to be higher than that usually used to distinguish the folded and unfolded states, to take into account the fact that the biological function of proteins may be disrupted by a slight structure deformation. The exact values of the above parameters are adjustable to some extent; however, their changes within a range only slightly shift the conformational phase boundaries, not affecting the major conclusions drawn from the results.
To measure the magnitude of the changes of secondary structures of the protein under the influence of the NP, the relative fraction of secondary structure is calculated by normalizing the number of contacts in the secondary structure when the NP is present with respect to the corresponding value when the NP is not, with the other conditions being the same. Therefore, a value smaller than 1 suggests a destabilized secondary structure, while a value larger than 1 suggests an enhanced one by the NPs.
2.2 Molecular Dynamics Simulations
The protein was modeled with an off-lattice C\(_\alpha \) based Go-type model; the residues were represented as beads centered at their C\(_\alpha \) positions and interacting with each other through bonds, angles, dihedral angles, and 12-10 Lennard-Jones interactions [22]. This protein model has been extensively used in studying protein folding and dynamics and has achieved enormous success [22,23,24,25,26].
The interactions between the protein residue i and NP were modeled by a Lennard-Jones potential,
if both have the same hydrophobicities or
if not, where  , \(\varepsilon _2=1.0\varepsilon \) and \(\varepsilon _3=\varepsilon _{npp}\varepsilon \), which determines the interaction strength between residues and the NP. \(r_i\) is the distance between the ith residue and the center of the NP, and D is the diameter of the NP.
, \(\varepsilon _2=1.0\varepsilon \) and \(\varepsilon _3=\varepsilon _{npp}\varepsilon \), which determines the interaction strength between residues and the NP. \(r_i\) is the distance between the ith residue and the center of the NP, and D is the diameter of the NP.
The native contacts within proteins were determined based on the PDB structures of the proteins. Specifically, a native contact between residues i and j (\(|i-j|>4\)) is defined if any distance between the heavy atoms (non-hydrogen) of two residues is smaller than a cutoff of  . In post processing the trajectories after the computations finished, a contact was deemed to be formed when any distance between heavy atoms is less than 1.2 times of its native value; this is to take account of the thermal fluctuations of the structures at finite temperatures. The fraction of the formed native contacts, denoted as Q, was used to measure the closeness of the structure to the native one and characterize the folding extents of the protein; its maximum value 1 indicates the structure is the same with the native one. The interplay between protein and NP was also characterized by their contacts, here a contact between protein and the NP was deemed to be formed if the distance between the center of the residue bead and the surface of the NP was less than 1.2\(d_0\).
. In post processing the trajectories after the computations finished, a contact was deemed to be formed when any distance between heavy atoms is less than 1.2 times of its native value; this is to take account of the thermal fluctuations of the structures at finite temperatures. The fraction of the formed native contacts, denoted as Q, was used to measure the closeness of the structure to the native one and characterize the folding extents of the protein; its maximum value 1 indicates the structure is the same with the native one. The interplay between protein and NP was also characterized by their contacts, here a contact between protein and the NP was deemed to be formed if the distance between the center of the residue bead and the surface of the NP was less than 1.2\(d_0\).
In all simulations, the protein was constrained in a finite spherical space with a soft wall, the radius of the space is set to 5 nm. The NP was fixed at the center of this space.
Molecular dynamics (MD) was employed to evolve the system. The equation of motion is described by a Langevin equation as follows [28].
where m is the mass of the bead, \(F_c=-\partial V / \partial x\) is the conformational force, \(\gamma \) is the friction coefficient, and \(\varGamma \) is the Gaussian random force, which is used to balance the energy dissipation caused by friction [28]. The random force satisfies the autocorrelation function
where \(k_B\) is the Boltzmann constant and T is the absolute temperature.
For each parameter combination, multiple long MD simulations (\({>}10^8\) MD steps) were carried out with each starting from different initial protein locations and at slightly different temperatures (around \(0.9T_f\), where \(T_f\) is the folding temperature of the protein) to avoid possible kinetic trapping. Weighted histogram analysis method (WHAM) was used to combine these multiple trajectories and reweight them to \(0.9T_f\) to calculate the thermodynamic properties [29,30,31].
3 Results and Discussion
3.1 Phase Diagram of \(\alpha +\beta \) Protein and Janus NP
Figure 1 shows the conformational phase diagram for protein GB1 adsorbed on a Janus NP whose surface is half hydrophobic and half hydrophilic. Here the term phase diagram is borrowed from statistical physics to describe the different conformational status of the protein-NP complex; it does not necessarily imply there is a change such as heat capacity associated with the phase transition. The conformation status (or phase) of the protein is simply divided into two categories, i.e. the folded and unfolded states, since only the general effects of NPs on proteins are of interest here. There are only two phases that can be identified in the diagram which are DF and AU. According to the diagram, if both the values of interaction strength \(\varepsilon _{npp}\) between protein and NP and the diameter D of NP are small (\(\varepsilon _{npp}<7\) and \(D/D_0<0.43\)), the protein is dissociated from NP and keeps folded (the DF region). On the contrary, if both the values of \(\varepsilon _{npp}\) and D are large enough (\(\varepsilon _{npp}>7.5\) and \(D/D_0>0.48\)), the protein is adsorbed on the NP without keeping its native structure (the AU region). Different from our previous work [32], the fact that the phase diagram has only two regions results from the effects of both hydrophobic and hydrophilic sides of the NP. On one hand, the hydrophobic surface may lead to the unfolding of the protein by attacking its hydrophobic core. On the other hand, the hydrophilic surface makes the protein spread around the NP surface, which also unfolds the protein. As a result, once the protein is adsorbed on the surface of the nanoparticles, it is strongly driven to unfold. An example can be seen in AU region in Fig. 2. The structure indicates that hydrophobic residues are more likely to be adsorbed by hydrophobic surface, while the hydrophilic residues are more likely to be adsorbed by hydrophilic surface. However, because of the hydrophobicity of sequence, some hydrophobic residues are adsorbed by hydrophilic surface and some hydrophilic residues are adsorbed by hydrophobic surface. This leads to the twist of the protein and the protein is in a loose bound to the NP. Thus, the binding of protein and Janus NP is always accompanied with the unfolding of protein.
Phase diagram for the adsorption and folding status of protein GB1 in close proximity to a Janus NP under different interaction strengthes (\(\varepsilon _{npp}\)) and NP diameters (D). The labels for the phases are explained in the Method section. The embedded structures provide direct pictures for the corresponding phases. The blue part of the protein chain in the structure indicates the hydrophobic residues, and the red part indicates the hydrophilic residues. The blue part of the nanoparticles represents the hydrophobic surface, and the red part represents the hydrophilic surface. (Color figure online)
The boundary of DF and AU regions are functions of both the interaction strength \(\varepsilon _{npp}\) and the diameter D. The larger \(\varepsilon _{npp}\) is, the smaller D is, and vice versa. This is due to the fact that the increase of interaction strength \(\varepsilon _{npp}\) leads to large interaction energy, meanwhile NP with large size has large surface energy [33], which may both result in the unfolding of the protein. Therefore, the two factors perform negative correlation at the boundary. The conformational phase diagram is different from our previous work where a protein adsorbed on NPs with a whole hydrophobic surface or a whole hydrophilic surface [32]. The former one has only two phases DF and AU, while the latter one has four phases DF, AF, AU and WU. The disappearance of the two phases AF and WU may due to the inconsistent distribution of the hydrophobic distribution of protein residues and the surface hydrophobicity of Janus NPs. This inconsistency makes the protein tend to unfold when it is adsorbed on the surface of Janus NPs.
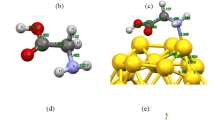
The representative structure of GB1 of the largest cluster, plotted for AU phase. The structures are plotted from two view directions that are perpendicular to each other ((a) and (b)), with the parameters \(\varepsilon _{npp}=7\varepsilon \) and  . The hydrophobic residues are in red and the hydrophilic residues in green. The hydrophobic surface of Janus NP is in blue and the hydrophilic surface in red. (Color figure online)
. The hydrophobic residues are in red and the hydrophilic residues in green. The hydrophobic surface of Janus NP is in blue and the hydrophilic surface in red. (Color figure online)
3.2 Binding Probability of Protein Residues on NP Surface
To further verify the above arguments, we calculated the binding probability of each residue upon the NP surface. As can be seen in Fig. 3, hydrophobic residues 5–7 incline to bind with the hydrophobic surface of the NP, while the residues 8–11 next to them are all hydrophilic ones and tend to bind with the hydrophilic half of the NP. Therefore, the structure is twisted to fit for the hydrophobicity. Similarly, the residues 31–42 have high probability to bind with the hydrophilic surface, since most of them are hydrophilic residues, while residues 30, 33 and 39 around them are hydrophobic ones and have high probability to bind with the hydrophobic surface. Therefore, the structure of this sequence has also been distorted. Besides, the overall binding probability of each residue is less than 0.6, which is much lower than the binding probability of protein adsorbed on NPs with a whole hydrophobic/hydrophilic surface (the maximum value reaches over 0.9 [32]). This suggests that the protein is loosely bound to the Janus NP, which can also be verified by the structures in Fig. 2. The loosely binding may also result from the disturbing of protein structure caused by the mismatch of hydrophobicity distributions between protein residues and the Janus NP.
The hydrophobicities of the residues and their binding probabilities on NPs. The first rod shows the hydrophobicity of the protein sequence, with blue denoting the hydrophobic residues and red otherwise. The second rod represents the binding probability of the corresponding residues and the hydrophobic surface of the Janus NP in the AU region, while the third one represents that of the hydrophilic surface. The deeper the color, the higher the binding probability. (Color figure online)
3.3 Transition Property of Binding Process
To gain further insights into the phase transition process, we calculated how the protein structure and the protein-NP contacting area S change from one phase to another at the phase boundary, as shown in Fig. 4; also shown are the typical trajectories collected at the corresponding phase boundary. In general, for a NP of fixed size (Fig. 4(a) and (b)), the Q value decreases and S increases when \(\varepsilon _{npp}\) is enlarged. This reflects the coupled adsorption and unfolding of the protein, which is consistent with the experiments that large contact areas decrease thermodynamic stabilities [8, 27]. Whereas, for a fixed interaction strength (Fig. 4(c) and (d)), the Q value gradually decreases with the increase of D, while the S value first increases and then decreases slightly after D is larger than a threshold, which is around  , almost the size of hydrophobic core of the protein. The transition caused by increasing D is also a cooperativity between unfolding and binding. In the DF region, although the protein is dissociated from the NP, the structure is still affected by the NP and the protein starts to spread on the NP, since they are very close to each other. The size of NP is smaller than the protein’s hydrophobic core, therefore, it cannot lead to significant structure change. In the AU region, the size of NP becomes larger than the hydrophobic core, and it makes the protein unfold with the bias to buring inside the protein. The decrease of relative contact area S value is due to that the surface area of NP increases significantly, while the contact area changes little since the protein is loosely bound to the NP, then the ratio decreases.
, almost the size of hydrophobic core of the protein. The transition caused by increasing D is also a cooperativity between unfolding and binding. In the DF region, although the protein is dissociated from the NP, the structure is still affected by the NP and the protein starts to spread on the NP, since they are very close to each other. The size of NP is smaller than the protein’s hydrophobic core, therefore, it cannot lead to significant structure change. In the AU region, the size of NP becomes larger than the hydrophobic core, and it makes the protein unfold with the bias to buring inside the protein. The decrease of relative contact area S value is due to that the surface area of NP increases significantly, while the contact area changes little since the protein is loosely bound to the NP, then the ratio decreases.
The coupled adsorption and unfolding of the protein as a function of the proteinCNP interaction strength \(\varepsilon _{npp}\) on crossing the phase boundary. Black curves with solid squares give the protein nativeness Q, and blue curves with open triangles show the fraction of NP surface covered by protein S. On the right side, typical trajectories collected right at the phase boundaries are shown. The parameters (\(\varepsilon _{npp}\), \(D/D_0\)) used to calculate the trajectories are (7, 0.58) and (8, 0.38) for (b) and (d), respectively. (Color figure online)
The relative fraction of \(\alpha \)-helix and \(\beta \)-sheets of protein GB1 when adsorbed on a Janus NP, normalised against their corresponding values obtained in simulations at the same conditions but without the NP. Error bars are shown as vertical line segments at the top of the histograms. (Color figure online)
3.4 Secondary Structures Affected by Janus NP
The presence of a nearby NP has different effects on different secondary structures of proteins. Figure 5 shows the fraction of secondary structures in each phase with respect to their values without the NP. The values are averaged over an equally spaced grid in the corresponding phase to reflect the general feature. If the value is smaller than 1, it indicates that the secondary structure is destabilized. If the value is larger than 1, it suggests that the secondary structure is strengthened due to the presence of NP. According to Fig. 5, in the DF phase, the fractions of hairpins \(\beta _{12}\) and \(\beta _{34}\) are increased compared with the fractions in the native structure, while the fraction of hairpin \(\beta _{14}\) is not sensitive to the presence of NP, as can be seen from the near-unity value. According to our previous study the instability of \(\beta \)-hairpin is not resulted from crowding effect [32]. In the DF region, the protein keeps folded and close to the NP. It seldom departs away from the NP, and the stability of hairpin structure is little affected by the space effect. The fraction of \(\alpha \)-helix decreases slightly, indicating the decrease of helical structure. In the DF phase, although the protein is not adsorbed on the NP, it is still weakly attached to the NP, and hence the protein is still under the influence of the NP. The enhancement of \(\beta \)-structures can be tentatively attributed to its geometry properties. \(\beta \)-hairpin is composed of inter-strand long-order hydrogen bonds, which makes it more flexible and tolerable to the curvature of the NP. The \(\alpha \)-helix is composed of inner-strand hydrogen bonds, which makes it more rigid and is easy to unfold when binding to NP. The above observations are consistent with the previous experiments that suggested that upon adsorption the protein underwent a change of secondary structure with a decrease of \(\alpha \)-helices and a possible increase of \(\beta \)-sheet structures [33, 34].
4 Conclusion
In summary, we studied how a \(\alpha \)+\(\beta \) protein would be affected by a nearby Janus NP with half hydrophobic surface and half hydrophilic surface using a structure-based coarse-grained model and molecular simulations. While most previous experimental and theoretical studies have focused on NPs of size of several 10 or 100 nm, our study is interested in ultra-small NPs less than several nanometres. According to our simulation results, the conformational phase diagrams show two phases, including DF and AU. The exact phase where the protein-NP system belongs to is dependent on the protein-NP interaction strength \(\varepsilon _{npp}\) and the NP size. In general, large NPs and strong interaction strengths denature proteins to large extents, consistent with previous theoretical and experimental studies. Our simulations also show that the NP exerts different effects on different secondary structures of the protein. Although in the unfolded phases both \(\alpha \)- and \(\beta \)-structures are destabilised, the \(\beta \)-structures are often enhanced in the folded phases and this enhancement is irrelevant to the hydrophobicity of the NP, the spatial organising pattern of the hydrophobic/hydrophilic residues or the crowding effect. This enhancement is tentatively attributed to the geometry and flexibility of the \(\beta \)-structures. The results may be useful for understanding the interaction of proteins with ultra-small NPs, particularly the fullerene derivatives and carbon nanotubes, considering the sizes of the NPs studied here. Our study illustrated all the possible structures of protein and Janus NP, and revealed the physical mechanism of the influence of NPs on secondary structures. This provides a deeper understanding of protein-NP system and is useful for the design of new materials.
The interactions between proteins and NPs are in fact very complex; in addition to the physical parameters considered in this work, they are also dependent on the protein and NP concentration, surface modifications of the NP, temperature, pH-value, denaturants in solvent, and etc. Further studies are ongoing in our group and they may deepen our understanding on such a system in different environments and facilitate designing new bio-nano-materials or devices.
References
Mahmoudi, M., Lynch, I., Ejtehadi, M.R., et al.: Protein-nanoparticle interactions: opportunities and challenges. Chem. Rev. 111, 5610–5637 (2011)
Sarikaya, M., Tamerler, C., Jen, A.K.Y., Schulten, K., Baneyx, F.: Molecular biomimetics: nanotechnology through biology. Nat. Mater. 2, 577–585 (2003)
Guo, X., Zhang, J., Wang, W.: The interactions between nanomaterials and biomolecules and the microstructural features. Prog. Phys. 32, 285–293 (2012)
Howes, P.D., Chandrawati, R., Stevens, M.M.: Bionanotechnology. Colloidal nanoparticles as advanced biological sensors. Science 346, 1247390 (2014)
Nair, R.R., Wu, H.A., Jayaram, P.N., Grigorieva, I.V., Geim, A.K.: Unimpeded permeation of water through helium-leak-tight graphene-based membranes. Science 335, 442 (2012)
Peng, Q., Mu, H.: The potential of protein-nanomaterial interaction for advanced drug delivery. J. Controlled Release Off. 225, 121 (2016)
Simunovic, M., Evergren, E., Golushko, I., Prvost, C., Renard, H.F., Johannes, L., et al.: How curvature-generating proteins build scaffolds on membrane nanotubes. Proc. Natl. Acad. Sci. USA 113, 11226 (2016)
Shang, W., Nuffer, J.H., Dordick, J.S., Siegel, R.W.: Unfolding of ribonuclease A on silica nanoparticle surfaces. Nano Lett. 7, 1991–1995 (2007)
Gagner, J.E., Lopez, M.D., Dordick, J.S., Siegel, R.W.: Effect of gold nanoparticle morphology on adsorbed protein structure and function. Biomaterials 32, 7241–7252 (2011)
Rocha, S., Thnemann, A.F., Pereira, M.C., et al.: Influence of fluorinated and hydrogenated nanoparticles on the structure and fibrillogenesis of amyloid beta-peptide. Biophys Chem. 137, 35–42 (2008)
Duong, H.T.T., Nguyen, D., Neto, C., Hawkett, B.S.: Synthesis and Applications of Polymeric Janus Nanoparticles: Synthesis, Self-Assembly, and Applications (2017)
Kobayashi, Y., Arai, N.: Self-assembly and viscosity behavior of Janus nanoparticles in nanotube flow. Langmuir 33, 736 (2017)
Lee, K., Yu, Y.: Janus nanoparticles for T cell activation: clustering ligands to enhance stimulation. J. Mater. Chem. B Mater. Biol. Med. 5, 4410–4415 (2017)
Li, F., Josephson, D.P., Stein, A.: Colloidal assembly: the road from particles to colloidal molecules and crystals. Angew. Chem. Int. Ed. 50, 360–388 (2011)
Yoshida, M., Roh, K.-H., Mandal, S., Bhaskar, S., Lim, D., Nandivada, H., et al.: Structurally controlled bio-hybrid materials based on unidirectional association of anisotropic microparticles with human endothelial cells. Adv. Mater. 21, 4920–4925 (2009)
Roh, K.H., Martin, D.C., Lahann, J.: Biphasic Janus particles with nanoscale anisotropy. Nat. Mater. 4, 759–763 (2005)
Honegger, T., Sarla, S., Lecarme, O., Berton, K., Nicolas, A., Peyrade, D.: Selective grafting of proteins on Janus particles: adsorption and covalent coupling strategies. Microelectron. Eng. 88, 1852–1855 (2011)
Xia, X.R., MonteiroRiviere, N.A., Riviere, J.E.: An index for characterization of nanomaterials in biological systems. Nat. Nanotechnol. 5, 671 (2010)
Nie, K., Wu, W.P., Zhang, X.L., Yang, S.M.: Molecular dynamics study on the grain size, temperature, and stress dependence of creep behavior in nanocrystalline nickel. J. Mat. Sci. 52, 2180–2191 (2017)
Vilhena, J.G., Rubio-Pereda, P., Vellosillo, P., Serena, P.A., Prez, R.: Albumin (BSA) adsorption over graphene in aqueous environment: influence of orientation, adsorption protocol, and solvent treatment. Langmuir 32, 1742 (2016)
Kharazian, B., Hadipour, N.L., Ejtehadi, M.R.: Understanding the nanoparticle-protein corona complexes using computational and experimental methods. Int. J. Biochem. Cell Biol. 75, 162 (2016)
Clementi, C., Nymeyer, H., Onuchic, J.N.: Topological and energetic factors: what determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? an investigation for small globular proteins. J. Mol. Biol. 298, 937 (2000)
Koga, N., Takada, S.: Roles of native topology and chain-length scaling in protein folding: a simulation study with a Go-like model. J. Mol. Biol. 313, 171–180 (2001)
Chan, H.S., Zhang, Z., Wallin, S., Liu, Z.: Cooperativity, local-nonlocal coupling, and nonnative interactions: principles of protein folding from coarse-grained models. Annu. Rev. Phys. Chem. 62, 301–326 (2011)
Wang, W., Xu, W.X., Levy, Y., et al.: Confinement effects on the kinetics and thermodynamics of protein dimerization. Proc. Natl. Acad. Sci. USA 106, 5517–5522 (2009)
Guo, X., Zhang, J., Chang, L., Wang, J., Wang, W.: Effectiveness of Phi-value analysis in the binding process of Arc repressor dimer. Phys. Rev. E 84, 011909 (2011)
Shang, W., Nuffer, J.H., et al.: Cytochrome C on silica nanoparticles: influence of nanoparticle size on protein structure, stability, and activity. Small 5, 470–476 (2009)
Veitshans, T., Klimov, D., Thirumalai, D.: Protein folding kinetics: timescales, pathways and energy landscapes in terms of sequence-dependent properties. Fold Des. 2, 1–22 (1997)
Ferrenberg, A.M., Swendsen, R.H.: New Monte Carlo technique for studying phase transitions. Phys. Rev. Lett. 61, 2635 (1988)
Ferrenberg, A.M., Swendsen, R.H.: Optimized Monte Carlo data analysis. Phys. Rev. Lett. 63, 1195 (1989)
Kumar, S., Rosenberg, J.M., Bouzida, D., Swendsen, R.H., Kollman, P.A.: The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 13, 1011–1021 (1992)
Guo, X., Wang, J., Zhang, J., Wang, W.: Conformational phase diagram for proteins absorbed on ultra-small nanoparticles studied by a structure-based coarse-grained model. Mol. Simul. 41, 1200–1211 (2015)
Vertegel, A.A., Siegel, R.W., Dordick, J.S.: Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir 20, 6800–6807 (2004)
Shang, L., Wang, Y., Jiang, J., Dong, S.: pH-dependent protein conformational changes in albumin: gold nanoparticle bioconjugates: a spectroscopic study. Langmuir 23, 2714–2721 (2007)
Acknowledgements
This work was supported by Natural science fund for colleges and universities in Jiangsu Province (No. 16KJB140014, 16KJB180007, 17KJD510006), NSFC (No. 11504043), Scientific fund (No: 201710929002, 2011QDZR-11).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Guo, X., Zhao, X., Fang, S., Bian, Y., Kang, W. (2018). Computing Simulation of Interactions Between \(\alpha \)+\(\beta \) Protein and Janus Nanoparticle. In: Shi, Y., et al. Computational Science – ICCS 2018. ICCS 2018. Lecture Notes in Computer Science(), vol 10862. Springer, Cham. https://doi.org/10.1007/978-3-319-93713-7_34
Download citation
DOI: https://doi.org/10.1007/978-3-319-93713-7_34
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-93712-0
Online ISBN: 978-3-319-93713-7
eBook Packages: Computer ScienceComputer Science (R0)