Abstract
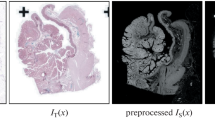
Registration of histology whole slide images of consecutive sections of a tissue block is mandatory for cross-slide analysis. Due to the stain variations, a feature-based method for deriving the transformation maps for these images is considered to be a reasonable choice as compared to the methods which work on image intensities. Autoencoders have been employed in a wide variety of applications due to their potential for representation learning and transfer learning for deep architectures. Representation learned by autoencoders has been used for a number of challenging problems including classification and regression. In this study, we analyze deep autoencoder features for the purpose of registering histology images by maximizing the feature similarities between the fixed and moving images. In this paper, we demonstrate the capability of autoencoder features for registration of histology images.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Zitova, B., Flusser, J.: Image registration methods: a survey. Image Vis. Comput. 21(11), 977–1000 (2003)
Moles Lopez, X., et al.: Registration of whole immunohistochemical slide images: an efficient way to characterize biomarker colocalization. J. Am. Med. Inf. Assoc. 22(1), 86–99 (2014)
Meyer, C.R., et al.: A methodology for registration of a histological slide and in vivo mri volume based on optimizing mutual information. Molecular imaging 5(1), 16–23 (2006)
Trahearn, N., Epstein, D., Snead, D., Cree, I., Rajpoot, N.: A fast method for approximate registration of whole-slide images of serial sections using local curvature. In: Medical Imaging 2014: Digital Pathology, vol. 9041, p. 90410E. International Society for Optics and Photonics (2014)
Trahearn, N., Epstein, D., Cree, I., Snead, D., Rajpoot, N.: Hyper-stain inspector: a framework for robust registration and localised co-expression analysis of multiple whole-slide images of serial histology sections. Sci. Rep. 7(1), 5641 (2017)
Mueller, D., Vossen, D., Hulsken, B.: Real-time deformable registration of multi-modal whole slides for digital pathology. Comput. Med. Imag. Graph. 35(7–8), 542–556 (2011)
Magee, D., et al.: Histopathology in 3D: From three-dimensional reconstruction to multi-stain and multi-modal analysis. J. Pathol. Inf. 6 (2015)
Wu, G., Kim, M., Wang, Q., Gao, Y., Liao, S., Shen, D.: Unsupervised deep feature learning for deformable registration of MR brain images. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013. LNCS, vol. 8150, pp. 649–656. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-40763-5_80
Wang, S., Kim, M., Wu, G., Shen, D.: Scalable high performance image registration framework by unsupervised deep feature representations learning. In: Deep Learning for Medical Image Analysis, pp. 245–269. Elsevier (2017)
Araújo, T., et al.: Classification of breast cancer histology images using convolutional neural networks. PloS One 12(6), e0177544 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Awan, R., Rajpoot, N. (2018). Deep Autoencoder Features for Registration of Histology Images. In: Nixon, M., Mahmoodi, S., Zwiggelaar, R. (eds) Medical Image Understanding and Analysis. MIUA 2018. Communications in Computer and Information Science, vol 894. Springer, Cham. https://doi.org/10.1007/978-3-319-95921-4_34
Download citation
DOI: https://doi.org/10.1007/978-3-319-95921-4_34
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-95920-7
Online ISBN: 978-3-319-95921-4
eBook Packages: Computer ScienceComputer Science (R0)