Abstract
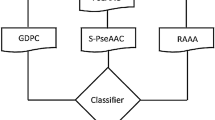
The research of the drug-target interactions (DTIs) is of great significance for drug development. Traditional chemical experiments are expensive and time-consuming. In recent years, many computational approaches based on different principles have been proposed gradually. Most of them use the information of drug-drug similarity and target-target similarity and made some progress. But the result is far from satisfactory. In this paper, we proposed machine learning method based on GBDT to predict DTIs with the IDs of both drug and protein, the descriptor of them, known DTIs and double negative samples. After gradient boosting and supervised training, GBDT construct decision trees for drug-target networks and generate precise model to predict new DTIs. Experimental results shows that Gradient Boosting Decision Tree (GBDT) reaches or outperforms other state-of-the-art methods.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Santos, R., Ursu, O., Gaulton, A., et al.: A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16(1), 19 (2017)
Law, V., Knox, C., Djoumbou, Y., et al.: DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 42(Database issue), D1091 (2014)
Wishart, D.S., Feunang, Y.D., Guo, A.C., et al.: DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46(D1), D1074–D1082 (2017)
Mei, J.P., Kwoh, C.K., Yang, P., et al.: Drugtarget interaction prediction by learning from local information and neighbors. Bioinformatics 29(2), 238–245 (2013)
DrugBank. https://www.drugbank.ca/
Van, L.T., Nabuurs, S.B., Marchiori, E.: Gaussian Interaction Profile Kernels for Predicting Drug-Target Interaction. Oxford University Press, Oxford (2011)
Van, L.T., Marchiori, E.: Predicting drug-target interactions for new drug compounds using a weighted nearest neighbor profile. PLoS One 8(6), e66952 (2013)
Gnen, M.: Predicting drugtarget interactions from chemical and genomic kernels using Bayesian matrix factorization. Bioinformatics 28(18), 2304 (2012)
Zheng, X., Ding, H., Mamitsuka, H., et al.: Collaborative matrix factorization with multiple similarities for predicting drug-target interactions. In: ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 1025–1033. ACM (2013)
Lu, Y., Guo, Y., Korhonen, A.: Link prediction in drug-target interactions network using similarity indices. BMC Bioinform. 18(1), 39 (2017)
Cao, D.S., Zhang, L.X., Tan, G.S., et al.: Computational prediction of drugtarget interactions using chemical, biological, and network features. Mol. Inform. 33(10), 669 (2014)
Cao, D.S., Liu, S., Xu, Q.S., et al.: Large-scale prediction of drug-target interactions using protein sequences and drug topological structures. Anal. Chim. Acta 752(21), 1 (2012)
Ding, Y., Tang, J., Guo, F.: Identification of drug-target interactions via multiple information integration. Inf. Sci. 418, 546–560 (2017)
Friedman, J.H.: Greedy function approximation: a gradient boosting machine. Ann. Stat. 29(5), 1189–1232 (2001)
Ye, J., Chow, J.H., Chen, J., et al.: Stochastic gradient boosted distributed decision trees. In: ACM Conference on Information and Knowledge Management, pp. 2061–2064 (2009)
Chen, T., He, T., Benesty, M.: Xgboost: extreme gradient boosting. R Package Version 0.4-2, 1–4 (2015)
Cao, D.S., Liang, Y.Z., Yan, J., et al.: PyDPI: freely available python package for chemoinformatics, bioinformatics, and chemogenomics studies. J. Chem. Inf. Model. 53(11), 3086–3096 (2013)
Lobo, J.M., Jimnez-Valverde, A., Real, R.: AUC: a misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 17(2), 145–151 (2008)
Rodriguez, J.D., Perez, A., Lozano, J.A.: Sensitivity analysis of k-fold cross validation in prediction error estimation. IEEE Trans. Pattern Anal. Mach. Intell. 32(3), 569–575 (2010)
Chen, X., Yan, C.C., Zhang, X., et al.: Drugtarget interaction prediction: databases, web servers and computational models. Brief. Bioinform. 17(4), 696 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Chen, J., Wang, J., Wang, X., Du, Y., Chang, H. (2018). Predicting Drug Target Interactions Based on GBDT. In: Perner, P. (eds) Machine Learning and Data Mining in Pattern Recognition. MLDM 2018. Lecture Notes in Computer Science(), vol 10934. Springer, Cham. https://doi.org/10.1007/978-3-319-96136-1_17
Download citation
DOI: https://doi.org/10.1007/978-3-319-96136-1_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-96135-4
Online ISBN: 978-3-319-96136-1
eBook Packages: Computer ScienceComputer Science (R0)