Abstract
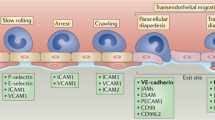
Cell-cell adhesion plays a critical role in the formation of tissues and organs. Adhesion between endothelial cells is also involved in the control of leukocyte migration across the endothelium of blood vessels. The most important players in this process are probably identified and the overall organization of the biochemical network can be drawn, but knowledge about connectivity is still incomplete, and the numerical values of kinetic parameters are unknown. This calls for qualitative modeling methods. Our aim in this paper is twofold: (i) to integrate in a unified model the biochemical network and the genetic circuitry. For this purpose we transform our system into a system of piecewise linear differential equations and then use Thomas theory of discrete networks. (ii) to show how constraints can be used to infer ranges of parameter values from observations and, with the same model, perform qualitative simulations.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Bockmayr, A., Courtois, A.: Using Hybrid Concurrent Constraint Programming to Model Dynamic Biological Systems. In: Stuckey, P.J. (ed.) ICLP 2002. LNCS, vol. 2401, pp. 85–99. Springer, Heidelberg (2002)
Chabrier, N., Fages, F.: Symbolic Model Checking of Biochemical Networks. In: Priami, C. (ed.) CMSB 2003. LNCS, vol. 2602, pp. 149–162. Springer, Heidelberg (2003)
Cohen, J.: Approaches for simulating and modeling cell regulation: search for a unified view using constraints. Linköping Electronic Articles in Computer and Information Science 3(7) (2001)
Colmerauer, A.: Prolog – Constraints Inside, Manuel de Prolog, PROLOGIA, Case 919, 13288 Marseille cedex 09, France (1996)
Delzanno, G., Podelski, A.: Model Checking in CLP. In: Cleaveland, W.R. (ed.) TACAS 1999. LNCS, vol. 1579, pp. 223–239. Springer, Heidelberg (1999)
Devloo, V., Hansen, P., Labbé, M.: Identification of All Steady States in Large Biological Systems by Logical Analysis. Bulletin of Mathematical Biology 65, 1025–1051 (2003)
Glass, L., Kauffman, S.A.: Co-operative components, spatial localization and oscillatory cellular dynamics. J. Theor. Biol. 34, 219–237 (1972)
Glass, L., Kauffman, S.A.: The logical analysis of continuous, non-linear biochemical control networks. J. Theor. Biol. 39, 103–129 (1973)
Gulino, D., Delachanal, E., Concord, E., Genoux, Y., Morand, B., Valiron, M.O., Sulpice, E., Scaife, R., Alemany, M., Vernet, T.: Alteration of Endothelial Cell Monolayer Integrity Triggers Resynthesis of Vascular Endothelium Cadherin. The Journal of Biological Chemistry 273, 29786–29793 (1998)
Hermant, B., Bibert, S., Concord, E., Dublet, B., Weidenhaupt, M., Vernet, T., Gulino-Debrac, D.: Identification of Proteases Involved in the Proteolysis of Vascular Endothelium Cadherin during Neutrophil Transmigration. The Journal of Biological Chemistry 278, 14002–14012 (2003)
Hickey, T.J., Wittenberg, D.K.: Rigorous Modeling of Hybrid Systems using Interval Arithmetic Constraints. In: Technical Report CS-03-241, Computer Science Departement, Brandeis University (2003)
Hordijk, P.L., Anthony, E., Mul, F.P., Rientsma, R., Oomen, L.C., Roos, D.: Vascular-Endothelial-Cadherin Modulates Endothelial Monolayer Permeability. J. Cell Sci. 112, 1915–1923 (1999)
de Jong, H., Gouzé, J.-L., Hernandez, C., Page, M., Sari, T., Geiselmann, J.: Hybrid modeling and simulation of genetic regulatory networks: A qualitative approach. In: Maler, O., Pnueli, A. (eds.) HSCC 2003. LNCS, vol. 2623, pp. 267–282. Springer, Heidelberg (2003)
Lampugnani, M.G., Corada, M., Caveda, L., Breviario, F., Ayalon, O., Geiger, B., Dejana, E.: The molecular organization of endothelial cell to cell junctions: differential association of plakoglobin, β-catenin, and α-catenin with vascular endothelial cadherin (VE-cadherin). J. Cell Biol. 129, 203–217 (1995)
Lampugnani, M.G., Resnati, M., Raiteri, M., Pigott, R., Pisacane, A., Houen, G., Ruco, L.P., Dejana, E.: A novel endothelial-specific membrane protein is a marker of cell-cell contacts. J. Cell Biol. 118, 1511–1522 (1992)
Legrand, P., Bibert, S., Jaquinod, M., Ebel, C., Hewatt, E., Vincent, F., Vanbelle, C., Concord, E., Vernet, T., Gulino, D.: Self-assembly of the vascular endothelial cadherin ectodomain in a Ca2 + -dependent hexameric structure. Journal of Biological Chemistry 276, 3581–3588 (2001)
Murray, J.D.: Mathematical Biology. Springer, Heidelberg (1989)
Peres, S., Comet, J.-P.: Contribution of Computational Tree Logic to Biological Regulatory Networks: Example from Pseudomonas Aeruginosa. In: Priami, C. (ed.) CMSB 2003. LNCS, vol. 2602, pp. 47–56. Springer, Heidelberg (2003)
Shapiro, B.E., Levchenko, A., Mjolsness, E.: Automatic model generation for signal transduction with applications to MAPK pathway. In: Kitano, H. (ed.) Foundations of Systems Biology. MIT Press, Cambridge (2002)
Snoussi, E.H., Thomas, R.: Logical Identification of All Steady States: The Concept of Feedback Loop Characteristic States. Bulletin of Mathematical Biology 55, 973–991 (1993)
Thieffry, D., Colet, M., Thomas, R.: Formalisation of Regulatory Networks: a Logical Method and Its Automation. Math. Modelling and Sci. Computing 2, 144–151 (1993)
Thomas, R., Kaufman, M.: Multistationarity, the Basis of Cell Differentiation and Memory. II. Logical Analysis of Regulatory Networks in Term of Feedback Circuits. Chaos 11, 180–195 (2001)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2005 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Fanchon, E., Corblin, F., Trilling, L., Hermant, B., Gulino, D. (2005). Modeling the Molecular Network Controlling Adhesion Between Human Endothelial Cells: Inference and Simulation Using Constraint Logic Programming. In: Danos, V., Schachter, V. (eds) Computational Methods in Systems Biology. CMSB 2004. Lecture Notes in Computer Science(), vol 3082. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-25974-9_9
Download citation
DOI: https://doi.org/10.1007/978-3-540-25974-9_9
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-25375-4
Online ISBN: 978-3-540-25974-9
eBook Packages: Computer ScienceComputer Science (R0)