Abstract
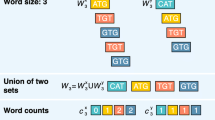
Given fragments from multiple genomes, we will show how to find an optimal local chain of colinear non-overlapping fragments in sub-quadratic time, using methods from computational geometry. A variant of the algorithm finds all significant local chains of colinear non-overlapping fragments. The local chaining algorithm can be used in a variety of problems in comparative genomics: The identification of regions of similarity (candidate regions of conserved synteny), the detection of genome rearrangements such as transpositions and inversions, and exon prediction.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Abouelhoda, M.I., Ohlebusch, E.: Multiple genome alignment: Chaining algorithms revisited. In: Baeza-Yates, R., Chávez, E., Crochemore, M. (eds.) CPM 2003. LNCS, vol. 2676, pp. 1–16. Springer, Heidelberg (2003)
Abouelhoda, M.I., Kurtz, S., Ohlebusch, E.: The enhanced suffix array and its applications to genome analysis. In: Guigó, R., Gusfield, D. (eds.) WABI 2002. LNCS, vol. 2452, pp. 449–463. Springer, Heidelberg (2002)
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J.: A basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990)
Batzoglou, S., Pachter, L., Mesirov, J.P., Berger, B., Lander, E.S.: Human and mouse gene structure: Comparative analysis and application to exon prediction. Genome Research 10, 950–958 (2001)
Bently, J.L.: K-d trees for semidynamic point sets. In: 6th Annual ACM Symposium on Computational Geometry, pp. 187–197. ACM, New York (1990)
Blattner, F.R., et al.: The complete genome sequence of Escherichia coli K-12. Science 277(5331), 1453–1474 (1997)
Bray, N., Dubchak, I., Pachter, L.: AVID: A global alignment program. Genome Research 13, 97–102 (2003)
Brudno, M., Morgenstern, B.: Fast and sensitive alignment of large genomic sequences. In: Proceedings of the IEEE Computer Society Bioinformatics Conference, pp. 138–150. IEEE, Los Alamitos (2002)
Chain, P., Kurtz, S., Ohlebusch, E., Slezak, T.: An applications-focused review of comparative genomics tools: Capabilities, limitations and future challenges. Briefings in Bioinformatics 4(2) (2003)
Chazelle, B.: A functional approach to data structures and its use in multidimensional searching. SIAM Journal on Computing 17(3), 427–462 (1988)
Delcher, A.L., Kasif, S., Fleischmann, R.D., Peterson, J., White, O., Salzberg, S.L.: Alignment of whole genomes. Nucleic Acids Res. 27, 2369–2376 (1999)
Delcher, A.L., Phillippy, A., Carlton, J., Salzberg, S.L.: Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30(11), 2478–2483 (2002)
Eisen, J.A., Heidelberg, J.F., White, O., Salzberg, S.L.: Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biology 1(6), 1–9 (2000)
Eppstein, D.: http://www.ics.uci.edu/eppstein/pubs/p-sparsedp.html
Eppstein, D., Giancarlo, R., Galil, Z., Italiano, G.F.: Sparse dynamic programming. I:Linear cost functions; II:Convex and concave cost functions. Journal of the ACM 39, 519–567 (1992)
Heidelberg, J.F., et al.: DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483 (2000)
Himmelreich, R., Plagens, H., Hilbert, H., Reiner, B., Herrmann, R.: Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 25, 701–712 (1997)
Hughes, D.: Evaluating genome dynamics: The constraints on rearrangements within bacterial genomes. Genome Biology 1(6), reviews 0006.1–0006.8 (2000)
Johnson, D.B.: A priority queue in which initialization and queue operations take O(log logD) time. Math. Sys. Theory 15, 295–309 (1982)
Kent, W.J., Zahler, A.M.: Conservation, regulation, synteny, and introns in a large-scale C.briggsae-C.elegans genomic alignment. Genome Research 10, 1115–1125 (2000)
Lee, D.T., Wong, C.K.: Worst-case analysis for region and partial region searches in multidimensional binary search trees and balanced quad trees. Acta Informatica 9, 23–29 (1977)
Höhl, M., Kurtz, S., Ohlebusch, E.: Efficient multiple genome alignment. In: Proceedings of the 10th International Conference on Intelligent Systems for Molecular Biology. Bioinformatics, vol. 18(Suppl. 1), pp. 312–320 (2002)
Morgenstern, B.: A space-efficient algorithm for aligning large genomic sequences. Bioinformatics 16, 948–949 (2000)
Myers, E.W., Miller, W.: Chaining multiple-alignment fragments in subquadratic time. In: Proceedings of the 6th ACM-SIAM Symposium on Discrete Algorithms, pp. 38–47 (1995)
Pearson, W.R., Lipman, D.J.: Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85, 2444–2448 (1988)
Preparata, F.P., Shamos, M.I.: Computational geometry: An introduction. Springer, New York (1985)
Roytberg, M.A., Ogurtsov, A.Y., Shabalina, S.A., Kondrashov, A.S.: A hierarchical approach to aligning collinear regions of genomes. Bioinformatics 18, 1673–1680 (2002)
Schwartz, S., Kent, J.K., Smit, A., Zhang, Z., Baertsch, R., Hardison, R., Haussler, D., Miller, W.: Human-mouse alignments with BLASTZ. Genome Research 13, 103–107 (2003)
Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., Miller, W.: PipMaker—A web server for aligning two genomic DNA sequences. Genome Research 10(4), 577–586 (2000)
van Emde Boas, P.: Preserving order in a forest in less than logarithmic time and linear space. Information Processing Letters 6(3), 80–82 (1977)
Vincens, P., Buffat, L., Andre, C., Chevrolat, J.P., Boisvieux, J.F., Hazout, S.: A strategy for finding regions of similarity in complete genome sequences. Bioinformatics 14, 715–725 (1998)
Zhang, Z., Raghavachari, B., Hardison, R.C., Miller, W.: Chaining multiplealignment blocks. J. Computational Biology 1, 51–64 (1994)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2003 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Abouelhoda, M.I., Ohlebusch, E. (2003). A Local Chaining Algorithm and Its Applications in Comparative Genomics. In: Benson, G., Page, R.D.M. (eds) Algorithms in Bioinformatics. WABI 2003. Lecture Notes in Computer Science(), vol 2812. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-39763-2_1
Download citation
DOI: https://doi.org/10.1007/978-3-540-39763-2_1
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-20076-5
Online ISBN: 978-3-540-39763-2
eBook Packages: Springer Book Archive