Abstract
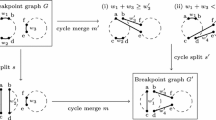
The genomic distance problem in the Hannenhalli-Pevzner theory is the following: Given two genomes whose chromosomes are linear, calculate the minimum number of inversions and translocations that transform one genome into the other. This paper presents a new distance formula based on a simple tree structure that captures all the delicate features of this problem in a unifying way.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Bergeron, A.: A very elementary presentation of the hannenhalli-pevzner theory. In: Amir, A., Landau, G.M. (eds.) CPM 2001. LNCS, vol. 2089, pp. 106–117. Springer, Heidelberg (2001)
Bergeron, A., Mixtacki, J., Stoye, J.: Reversal distance without hurdles and fortresses. In: Sahinalp, S.C., Muthukrishnan, S.M., Dogrusoz, U. (eds.) CPM 2004. LNCS, vol. 3109, pp. 388–399. Springer, Heidelberg (2004)
Bergeron, A., Mixtacki, J., Stoye, J.: On sorting by translocations. J. Comput. Biol. 13(2), 567–578 (2006)
Bergeron, A., Mixtacki, J., Stoye, J.: A unifying view of genome rearrangements. In: Bücher, P., Moret, B.M.E. (eds.) WABI 2006. LNCS (LNBI), vol. 4175, pp. 163–173. Springer, Heidelberg (2006)
Bergeron, A., Stoye, J.: On the similarity of sets of permutations and its applications to genome comparison. In: Warnow, T., Zhu, B. (eds.) COCOON 2003. LNCS, vol. 2697, pp. 68–79. Springer, Heidelberg (2003)
Hannenhalli, S., Pevzner, P.A.: Transforming men into mice (polynomial algorithm for genomic distance problem). In: Proceedings of FOCS 1995, pp. 581–592. IEEE Press, Los Alamitos (1995)
Jean, G., Nikolski, M.: Genome rearrangements: a correct algorithm for optimal capping. Inf. Process. Lett. 104, 14–20 (2007)
Ozery-Flato, M., Shamir, R.: Two notes on genome rearrangements. J. Bioinf. Comput. Biol. 1(1), 71–94 (2003)
Tesler, G.: Efficient algorithms for multichromosomal genome rearrangements. J. Comput. Syst. Sci. 65(3), 587–609 (2002)
Tesler, G.: GRIMM: Genome rearrangements web server. Bioinformatics 18(3), 492–493 (2002)
Yancopoulos, S., Attie, O., Friedberg, R.: Efficient sorting of genomic permutations by translocation, inversion and block interchange. Bioinformatics 21(16), 3340–3346 (2005)
Author information
Authors and Affiliations
Editor information
Rights and permissions
Copyright information
© 2008 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Bergeron, A., Mixtacki, J., Stoye, J. (2008). HP Distance Via Double Cut and Join Distance. In: Ferragina, P., Landau, G.M. (eds) Combinatorial Pattern Matching. CPM 2008. Lecture Notes in Computer Science, vol 5029. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-69068-9_8
Download citation
DOI: https://doi.org/10.1007/978-3-540-69068-9_8
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-69066-5
Online ISBN: 978-3-540-69068-9
eBook Packages: Computer ScienceComputer Science (R0)