Abstract
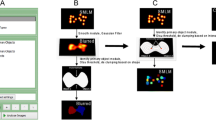
Recently, image-based, high throughput genome-wide RNA interference (RNAi) experiments are increasingly carried out to facilitate the understanding of gene functions in intricate biological processes. Effective automated segmentation technique is significant in analysis of RNAi images. Traditional graph cuts based active contours (GCBAC) method is impractical in automated segmentation. Here, we present a modified GCBAC approach to overcome this shortcoming. The whole process is implemented as follows: First, extracted nuclei are used in region-growing algorithm to get the initial contours for segmentation of cytoplasm. Second, constraint factor obtained from rough segmentation is incorporated to improve the performance of segmenting shapes of cytoplasm. Then, control points are searched to correct inaccurate parts of segmentation. Finally, morphological thinning algorithm is implemented to solve the touching problem of clustered cells. Our approach is capable of automatically segmenting clustered cells with low time-consuming. The excellent results verify the effectiveness of the proposed approach.
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Rafael, C.G., Richard, E.W.: Digital Image Processing, 2nd edn. Prentice-Hall, Englewood Cliffs (2002)
Kass, M., Witkin, A., Terzopoulos, D.: Snakes: Active contour models. Int. J. Comput. Vis. 1, 321–331 (1987)
Caselles, V., Kimmel, R., Sapiro, G.: Geodesic active contours. In: Proc. 5th International conf. on Computer Vision, Boston, pp. 694–699 (1995)
Xu, N., Bansal, R., Ahuja, N.: Object segmentation using graph cuts based active contours. In: CVPR 2001 Technical Sketches, pp. 87–90 (2001)
Ford, L., Fulkerson, D.: Flows in Networks. Princeton University Press, Princeton (1962)
Loupias, E., Sebe, N., Bres, S., Jolion: Wavelet-based salient points for image retrieval. In: Internat. Conf. on Image Processing, J.-M., vol. 2, pp. 518–521 (2000)
Otsu, N.: A threshold selection method from gray level histogram. IEEE Transactions on System man Cybernetics 8, 62–66 (1978)
Xiong, G.L., Zhou, X.B., Ji, L., Bradley, P., Perrimon, N., Wong, S.T.C.: Segmentation of drosophila RNAi fluorescence images using level sets. In: International Conference on Image Processing, Altanta, GA, USA (2006)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2006 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Chen, C., Li, H., Zhou, X. (2006). Automated Segmentation of Drosophila RNAi Fluorescence Cellular Images Using Graph Cuts. In: Cham, TJ., Cai, J., Dorai, C., Rajan, D., Chua, TS., Chia, LT. (eds) Advances in Multimedia Modeling. MMM 2007. Lecture Notes in Computer Science, vol 4351. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-69423-6_12
Download citation
DOI: https://doi.org/10.1007/978-3-540-69423-6_12
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-69421-2
Online ISBN: 978-3-540-69423-6
eBook Packages: Computer ScienceComputer Science (R0)